8JV4
 
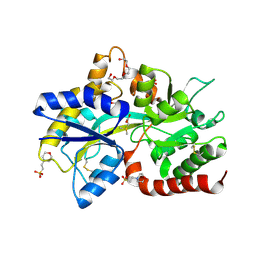 | | Structure of the SAR11 PotD in complex with DMSP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, GLYCEROL, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Structure of the SAR11 PotD in complex with DMSP
To Be Published
|
|
8JV3
 
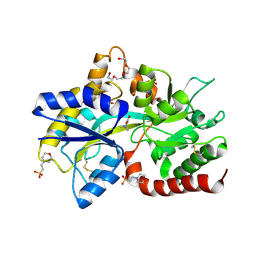 | |
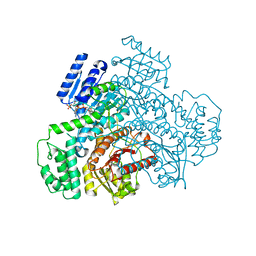
8J2D
 
 | |
7YMN
 
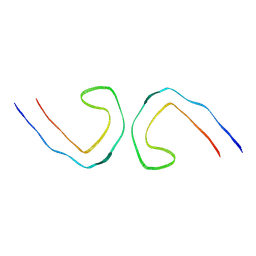 | | Cryo-EM structure of in vitro PHF fibril | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Li, X, Liu, C. | | Deposit date: | 2022-07-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Subtle change of fibrillation condition leads to substantial alteration of recombinant Tau fibril structure.
Iscience, 25, 2022
|
|
7YPG
 
 | |
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | Descriptor: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
6JKN
 
 | | Crystal Structure of G-quadruplex Formed by Bromo-substituted Human Telomeric DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*(BGM)P*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*(BGM)P*G)-3'), POTASSIUM ION | | Authors: | Geng, Y, Cai, Q, Liu, C, Zhu, G. | | Deposit date: | 2019-03-01 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | The crystal structure of an antiparallel chair-type G-quadruplex formed by Bromo-substituted human telomeric DNA.
Nucleic Acids Res., 47, 2019
|
|
6JKZ
 
 | | Crystal structure of VvPlpA from Vibrio vulnificus | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CHLORIDE ION, Thermolabile hemolysin | | Authors: | Ma, Q, Wan, Y, Liu, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-05-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Structural analysis of aVibriophospholipase reveals an unusual Ser-His-chloride catalytic triad.
J.Biol.Chem., 294, 2019
|
|
6JL1
 
 | |
6JL2
 
 | | Crystal structure of VvPlpA G389N from Vibrio vulnificus | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, HEXAETHYLENE GLYCOL, Thermolabile hemolysin | | Authors: | Ma, Q, Wan, Y, Liu, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of aVibriophospholipase reveals an unusual Ser-His-chloride catalytic triad.
J.Biol.Chem., 294, 2019
|
|
6K7Q
 
 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
6K7R
 
 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
6KJ4
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ1
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KZ4
 
 | | YebT domain 5-7 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Wang, H.W, Liu, C, Zhang, L. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM Structure of a Bacterial Lipid Transporter YebT.
J.Mol.Biol., 432, 2020
|
|
6KZ3
 
 | | YebT domain1-4 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Wang, H.W, Liu, C, Zhang, L. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structure of a Bacterial Lipid Transporter YebT.
J.Mol.Biol., 432, 2020
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
6LNI
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2020-06-24 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6K7S
 
 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, TOMUDEX, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
7YK2
 
 | | Cryo-EM structure of Apo-alpha-syn fibril | | Descriptor: | Alpha-synuclein | | Authors: | Xu, Q.H, Xia, W.C, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational Dynamics of an alpha-Synuclein Fibril upon Receptor Binding Revealed by Insensitive Nuclei Enhanced by Polarization Transfer-Based Solid-State Nuclear Magnetic Resonance and Cryo-Electron Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7YK8
 
 | | Cryo-EM structure of dLAG3-alpha-syn fibril | | Descriptor: | Alpha-synuclein | | Authors: | Xu, Q.H, Xia, W.C, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational Dynamics of an alpha-Synuclein Fibril upon Receptor Binding Revealed by Insensitive Nuclei Enhanced by Polarization Transfer-Based Solid-State Nuclear Magnetic Resonance and Cryo-Electron Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7VZF
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
7E0F
 
 | |
