2AHE
 
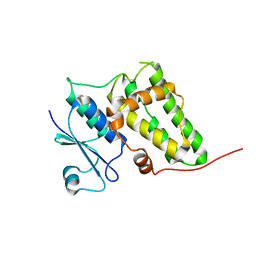 | | Crystal structure of a soluble form of CLIC4. intercellular chloride ion channel | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Littler, D.R, Assaad, N.N, Harrop, S.J, Brown, L.J, Pankhurst, G.J, Luciani, P, Aguilar, M.-I, Mazzanti, M, Berryman, M.A, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4.
FEBS J., 272, 2005
|
|
7N3K
 
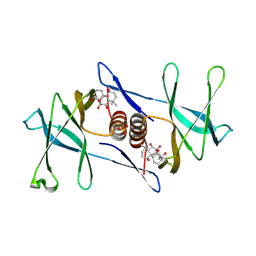 | | Oridonin-bound SARS-CoV-2 Nsp9 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A natural product compound inhibits coronaviral replication in vitro by binding to the conserved Nsp9 SARS-CoV-2 protein.
J.Biol.Chem., 297, 2021
|
|
8SHI
 
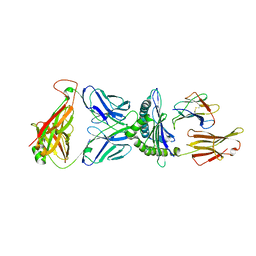 | | Valpha3S1 Vbeta13S1 HLA C 0602 VRSRRCLRL | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), T cell receptor alpha, ... | | Authors: | Littler, D.R, Anand, S, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.90001726 Å) | | Cite: | Complimentary electrostatics dominate T-cell receptor binding to a psoriasis-associated peptide antigen presented by human leukocyte antigen C∗06:02.
J.Biol.Chem., 299, 2023
|
|
3KJY
 
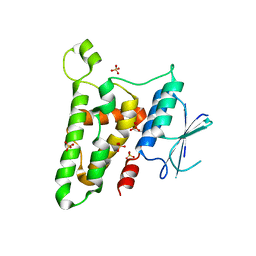 | | Crystal structure of reduced HOMO SAPIENS CLIC3 | | Descriptor: | Chloride intracellular channel protein 3, SULFATE ION | | Authors: | Littler, D.R, Curmi, P.M.G, Breit, S.N, Perrakis, A. | | Deposit date: | 2009-11-04 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human CLIC3 at 2 A resolution
Proteins, 78, 2010
|
|
7SKI
 
 | | Pertussis toxin in complex with PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.09912443 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7SKY
 
 | | Pertussis toxin S1 bound to NAD+ | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37000132 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7SKK
 
 | | pertussis toxin in complex with ADPR and Nicotinamide | | Descriptor: | NICOTINAMIDE, Pertussis toxin subunit 1, SULFATE ION, ... | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65000772 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
8DQU
 
 | | Nanobody bound SARS-CoV-2 Nsp9 | | Descriptor: | Nanobody, Non-structural protein 9 | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45003176 Å) | | Cite: | Inside-out: Antibody-binding reveals potential folding hinge-points within the SARS-CoV-2 replication co-factor nsp9.
Plos One, 18, 2023
|
|
8TUH
 
 | | HLA B7:02 with RPIIRPATL | | Descriptor: | 1,2-ETHANEDIOL, ARG-PRO-ILE-ILE-ARG-PRO-ALA-THR-LEU, Beta-2-microglobulin, ... | | Authors: | Littler, D.R, Rossjohn, J, Chaurasia, P, Petersen, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.200106 Å) | | Cite: | CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Nat Commun, 15, 2024
|
|
8TUB
 
 | | HLA B7:02 with HPNGYKSLSTL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Littler, D.R, Rossjohn, J, Chaurasia, P, Petersen, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Nat Commun, 15, 2024
|
|
5KBF
 
 | | cAMP bound PfPKA-R (141-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit, putative | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
5K8S
 
 | | cAMP bound PfPKA-R (297-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
6WXD
 
 | | SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
6WC1
 
 | |
6W9Q
 
 | | Peptide-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 3C-like proteinase peptide, Non-structural protein 9 fusion, PHOSPHATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
7SNE
 
 | | Pertussis toxin S1 subunit bound to BaAD | | Descriptor: | Pertussis toxin subunit 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylanilino)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Littler, D.R, Beddoe, T. | | Deposit date: | 2021-10-28 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.00011 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7U6Z
 
 | | Pertussis toxin E129D NAD | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.30002 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
2H6D
 
 | | Protein Kinase Domain of the Human 5'-AMP-activated protein kinase catalytic subunit alpha-2 (AMPK alpha-2 chain) | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Littler, D.R, Walker, J.R, Wybenga-Groot, L, Newman, E.M, Butler-Cole, C, Mackenzie, F, Finerty, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-31 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved mechanism of autoinhibition for the AMPK kinase domain: ATP-binding site and catalytic loop refolding as a means of regulation.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
8EMG
 
 | |
8EMF
 
 | |
8EO8
 
 | |
8EN8
 
 | |
8ENH
 
 | |
8EMK
 
 | |
8EMJ
 
 | |
