1B7G
 
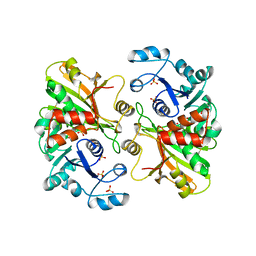 | | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE | | Descriptor: | PROTEIN (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Isupov, M.N, Littlechild, J.A. | | Deposit date: | 1999-01-22 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J.Mol.Biol., 291, 1999
|
|
5FIP
 
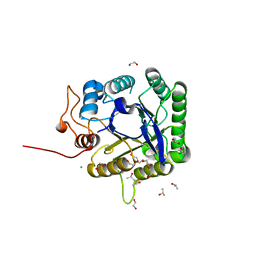 | | Discovery and characterization of a novel thermostable and highly halotolerant GH5 cellulase from an Icelandic hot spring isolate | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, ... | | Authors: | Zarafeta, D, Kissas, D, Sayer, C, Gudbergsdottir, S.R, Ladoukakis, E, Isupov, M.N, Chatziioannou, A, Peng, X, Littlechild, J.A, Skretas, G, Kolisis, F.N. | | Deposit date: | 2015-10-01 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Characterization of a Thermostable and Highly Halotolerant Gh5 Cellulase from an Icelandic Hot Spring Isolate.
Plos One, 11, 2016
|
|
5FRD
 
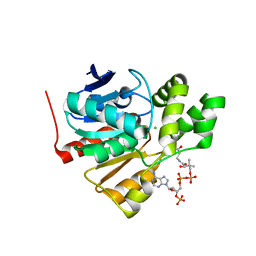 | | Structure of a thermophilic esterase | | Descriptor: | CARBOXYLESTERASE (EST-2), CHLORIDE ION, CITRATE ANION, ... | | Authors: | Sayer, C, Finnigan, W, Isupov, M.N, Levisson, M, Kengen, S.W.M, van der Oost, J, Harmer, N, Littlechild, J.A. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Characterisation of Archaeoglobus Fulgidus Esterase Reveals a Bound Coa Molecule in the Vicinity of the Active Site.
Sci.Rep., 6, 2016
|
|
1H2B
 
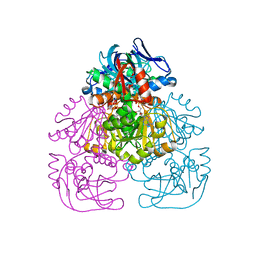 | | Crystal Structure of the Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Aeropyrum pernix at 1.65A Resolution | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Guy, J.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2002-08-02 | | Release date: | 2003-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
J.Mol.Biol., 331, 2003
|
|
3HGJ
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 complexed with p-hydroxy-benzaldehyde | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
2VIM
 
 | | X-ray structure of Fasciola hepatica thioredoxin | | Descriptor: | THIOREDOXIN | | Authors: | Line, K, Isupov, M.N, Garcia-Rodriguez, E, Maggioli, G, Parra, F, Littlechild, J.A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The Fasciola Hepatica Thioredoxin: High Resolution Structure Reveals Two Oxidation States.
Mol.Biochem.Parasitol., 161, 2008
|
|
3HF3
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
2VXI
 
 | | The binding of heme and zinc in Escherichia coli Bacterioferritin | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Willies, S.C, Isupov, M.N, Garman, E.F, Littlechild, J.A. | | Deposit date: | 2008-07-04 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Binding of Haem and Zinc in the 1.9 A X-Ray Structure of Escherichia Coli Bacterioferritin.
J.Biol.Inorg.Chem., 14, 2009
|
|
1HL7
 
 | | Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one | | Descriptor: | 3A,4,7,7A-TETRAHYDRO-BENZO [1,3] DIOXOL-2-ONE, GAMMA LACTAMASE | | Authors: | Line, K, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2003-03-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of a (-)Gamma-Lactamase from an Aureobacterium Species Reveals a Tetrahedral Intermediate in the Active Site
J.Mol.Biol., 338, 2004
|
|
1HKH
 
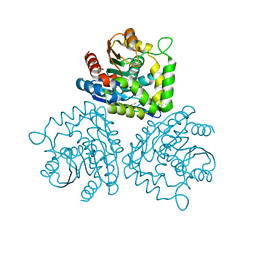 | |
6T94
 
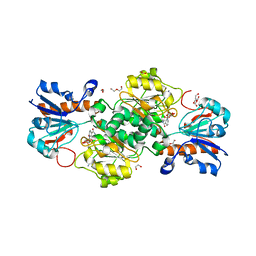 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T92
 
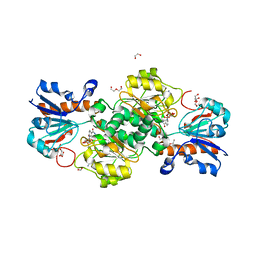 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Z
 
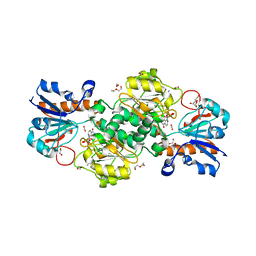 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
1QGD
 
 | |
5MR0
 
 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5NFQ
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5NG7
 
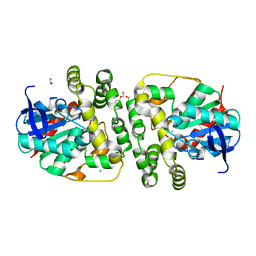 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
7QON
 
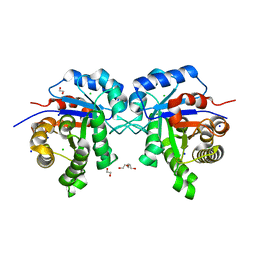 | |
1QHF
 
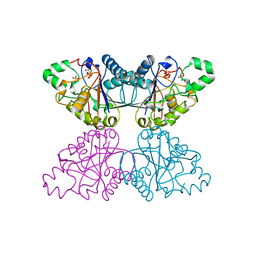 | | YEAST PHOSPHOGLYCERATE MUTASE-3PG COMPLEX STRUCTURE TO 1.7 A | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PROTEIN (PHOSPHOGLYCERATE MUTASE), SULFATE ION | | Authors: | Crowhurst, G, Littlechild, J, Watson, H.C. | | Deposit date: | 1999-05-13 | | Release date: | 1999-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QHB
 
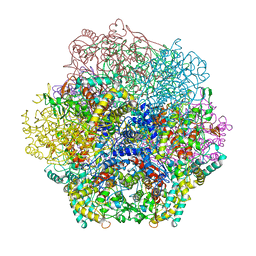 | | VANADIUM BROMOPEROXIDASE FROM RED ALGA CORALLINA OFFICINALIS | | Descriptor: | CALCIUM ION, HALOPEROXIDASE, PHOSPHATE ION | | Authors: | Isupov, M.N, Dalby, A.R, Brindley, A.A, Littlechild, J.A. | | Deposit date: | 1999-05-11 | | Release date: | 2000-07-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of dodecameric vanadium-dependent bromoperoxidase from the red algae Corallina officinalis.
J.Mol.Biol., 299, 2000
|
|
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | Descriptor: | PEROXIREDOXIN-2 | | Authors: | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | Deposit date: | 1999-10-07 | | Release date: | 2000-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
1QLW
 
 | |
4UHH
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (cacodylate complex) | | Descriptor: | CACODYLIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
