5ZGG
 
 | | NMR structure of p75NTR transmembrane domain in complex with NSC49652 | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(pyridin-3-yl)prop-2-en-1-one, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Targeting the Transmembrane Domain of Death Receptor p75NTRInduces Melanoma Cell Death and Reduces Tumor Growth.
Cell Chem Biol, 25, 2018
|
|
5XME
 
 | | Solution structure of C-terminal domain of TRADD | | Descriptor: | Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2017-05-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily.
Sci Rep, 7, 2017
|
|
5FBY
 
 | | Crystal structure of ctSPD | | Descriptor: | cleaved peptide, separase | | Authors: | Lin, Z, Luo, X, Yu, H. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structural basis of cohesin cleavage by separase.
Nature, 532, 2016
|
|
1YHP
 
 | | Solution Structure of Ca2+-free DdCAD-1 | | Descriptor: | Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
5CQR
 
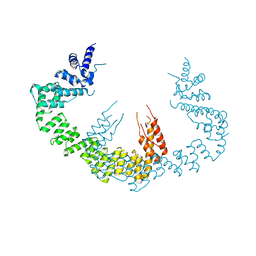 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQS
 
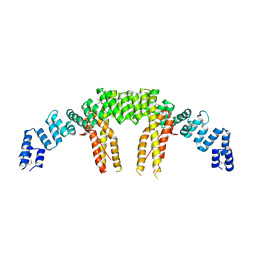 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6J68
 
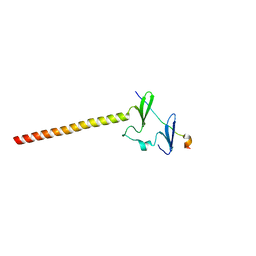 | | Structure of KIBRA and LATS1 Complex | | Descriptor: | Peptide from Serine/threonine-protein kinase LATS1, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JJY
 
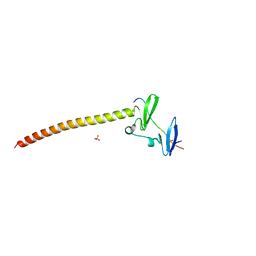 | | Crystal Structure of KIBRA and beta-Dystroglycan | | Descriptor: | Peptide from Dystroglycan, Protein KIBRA, SULFATE ION | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JJZ
 
 | | Crystal Structure of MAGI2 and Dendrin complex | | Descriptor: | ACETATE ION, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Lin, Z, Zhang, H, Yang, Z, Ji, Z, Zhang, M, Zhu, J. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JJX
 
 | | Crystal Structure of KIBRA and Angiomotin complex | | Descriptor: | Peptide from Angiomotin, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JK0
 
 | | Crystal Structure of YAP1 and Dendrin complex | | Descriptor: | CALCIUM ION, Transcriptional coactivator YAP1,Dendrin | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6J69
 
 | | Structure of KIBRA and Dendrin Complex | | Descriptor: | Peptide from Dendrin, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Kibra Modulates Learning and Memory via Binding to Dendrin.
Cell Rep, 26, 2019
|
|
6JJW
 
 | | Crystal Structure of KIBRA and PTPN14 complex | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JK1
 
 | | Crystal Structure of YAP1 and Dendrin complex 2 | | Descriptor: | Dendrin,Transcriptional coactivator YAP1 | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
7YGL
 
 | |
7YHL
 
 | |
7YGH
 
 | |
8X8T
 
 | |
3PB1
 
 | | Crystal Structure of a Michaelis Complex between Plasminogen Activator Inhibitor-1 and Urokinase-type Plasminogen Activator | | Descriptor: | Plasminogen activator inhibitor 1, Plasminogen activator, urokinase, ... | | Authors: | Lin, Z, Jiang, L, Huang, M, Structure 2 Function Project (S2F) | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1.
J.Biol.Chem., 286, 2011
|
|
2K3O
 
 | |
2K3N
 
 | |
2K3Q
 
 | |
2K3P
 
 | |
6L3F
 
 | |
6L8K
 
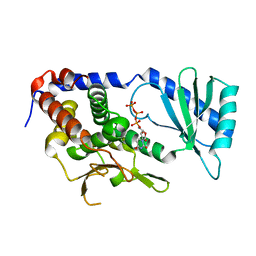 | | Structure of URT1 in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, UTP:RNA uridylyltransferase 1 | | Authors: | Lingru, Z. | | Deposit date: | 2019-11-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of Arabidopsis terminal uridylyl transferase URT1.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
