8WMM
 
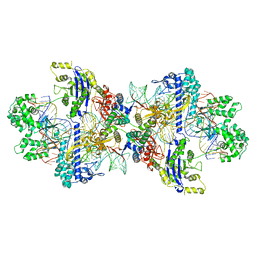 | | Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary) | | Descriptor: | MAGNESIUM ION, NTS, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WR4
 
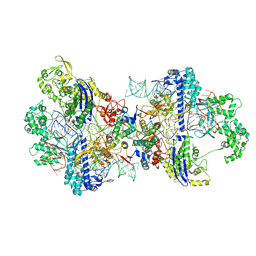 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
5GI7
 
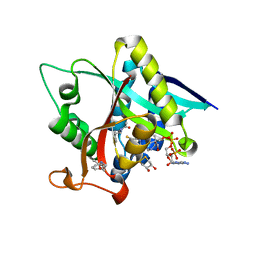 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-phenylethylamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
5GI9
 
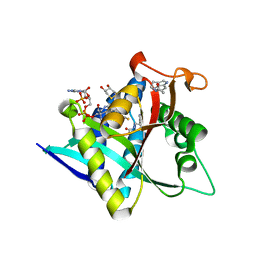 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-Tryptamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
7EPC
 
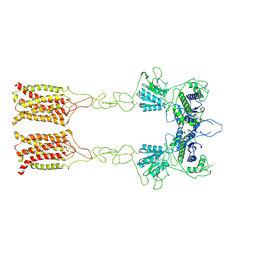 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
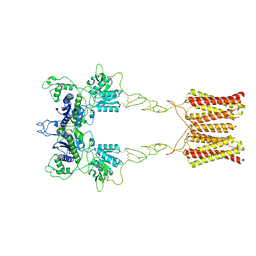 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPF
 
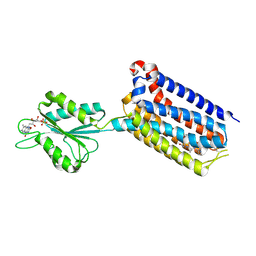 | | Crystal structure of mGlu2 bound to NAM597 | | Descriptor: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPB
 
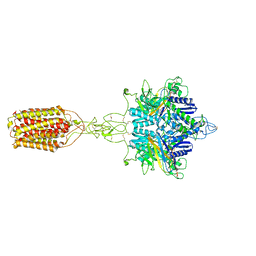 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
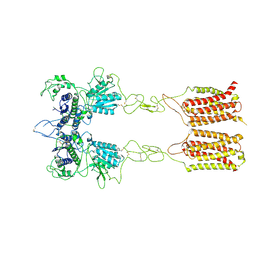 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7DYD
 
 | |
4BFN
 
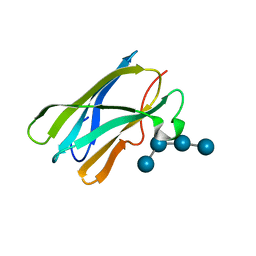 | | Crystal Structure of the Starch-Binding Domain from Rhizopus oryzae Glucoamylase in Complex with isomaltotetraose | | Descriptor: | GLUCOAMYLASE, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Chu, C.H, Li, K.M, Lin, S.W, Sun, Y.J. | | Deposit date: | 2013-03-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structures of Starch Binding Domain from Rhizopus Oryzae Glucoamylase in Complex with Isomaltooligosaccharide: Insights Into Polysaccharide Binding Mechanism of Cbm21 Family.
Proteins, 82, 2014
|
|
4BFO
 
 | | Crystal Structure of the Starch-Binding Domain from Rhizopus oryzae Glucoamylase in Complex with isomaltotriose | | Descriptor: | GLUCOAMYLASE, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Chu, C.H, Li, K.M, Lin, S.W, Sun, Y.J. | | Deposit date: | 2013-03-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.175 Å) | | Cite: | Crystal Structures of Starch Binding Domain from Rhizopus Oryzae Glucoamylase in Complex with Isomaltooligosaccharide: Insights Into Polysaccharide Binding Mechanism of Cbm21 Family.
Proteins, 82, 2014
|
|
1JTV
 
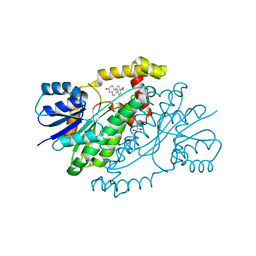 | | Crystal structure of 17beta-Hydroxysteroid Dehydrogenase Type 1 complexed with Testosterone | | Descriptor: | 17 beta-hydroxysteroid dehydrogenase type 1, GLYCEROL, TESTOSTERONE | | Authors: | Shi, R, Nahoum, V, Lin, S.X. | | Deposit date: | 2001-08-22 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Pseudo-symmetry of C19 steroids, alternative binding orientations, and
multispecificity in human estrogenic 17beta-hydroxysteroid
dehydrogenase.
FASEB J., 17, 2003
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | Descriptor: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
6VPV
 
 | | Trimeric Photosystem I from the High-Light Tolerant Cyanobacteria Cyanobacterium Aponinum | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Dobson, Z, Toporik, H, Vaughn, N, Lin, S, Williams, D, Fromme, P, Mazor, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structure of photosystem I from a high-light tolerant Cyanobacteria.
Elife, 10, 2021
|
|
1DHT
 
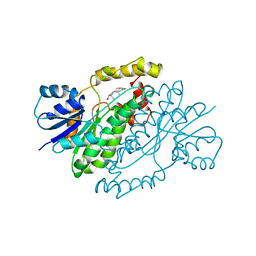 | | ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE COMPLEXED DIHYDROTESTOSTERONE | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE | | Authors: | Han, Q, Campbell, R.L, Gangloff, A, Lin, S.X. | | Deposit date: | 1998-03-25 | | Release date: | 1999-09-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Dehydroepiandrosterone and dihydrotestosterone recognition by human estrogenic 17beta-hydroxysteroid dehydrogenase. C-18/c-19 steroid discrimination and enzyme-induced strain.
J.Biol.Chem., 275, 2000
|
|
2VLC
 
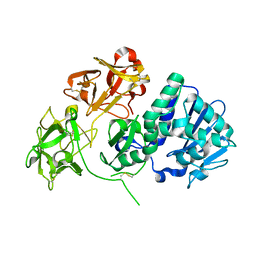 | | Crystal structure of Natural Cinnamomin (Isoform III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein, beta-D-mannopyranose, ... | | Authors: | Azzi, A, Wang, T, Zhu, D.-W, Zou, Y.-S, Liu, W.-Y, Lin, S.-X. | | Deposit date: | 2008-01-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Native Cinnamomin Isoform III and its Comparison with Other Ribosome Inactivating Proteins.
Proteins: Struct., Funct., Bioinf., 74, 2009
|
|
4CMQ
 
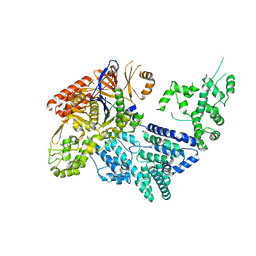 | | Crystal structure of Mn-bound S.pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MANGANESE (II) ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA- Mediated Conformational Activation
Science, 343, 2014
|
|
4CMP
 
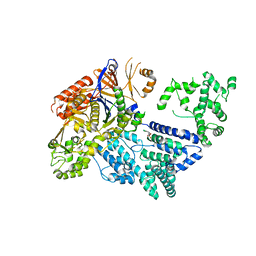 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
1MVE
 
 | | Crystal structure of a natural circularly-permutated jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes | | Descriptor: | CALCIUM ION, Truncated 1,3-1,4-beta-D-glucanase | | Authors: | Tsai, L.-C, Shyur, L.-F, Lee, S.-H, Lin, S.-S, Yuan, H.S. | | Deposit date: | 2002-09-25 | | Release date: | 2003-07-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Natural Circularly Permuted Jellyroll Protein: 1,3-1,4-beta-D-Glucanase from Fibrobacter succinogenes.
J.Mol.Biol., 330, 2003
|
|
1OV4
 
 | | Crystal structure of human DHEA-ST complexed with androsterone | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, Alcohol sulfotransferase, SULFATE ION | | Authors: | Chang, H.J, Shi, R, Rhese, P, Lin, S.X. | | Deposit date: | 2003-03-25 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identifying androsterone (ADT) as a cognate substrate for human dehydroepiandrosterone sulfotransferase (DHEA-ST) important for steroid homeostasis: structure of the enzyme-ADT complex.
J.Biol.Chem., 279, 2004
|
|
6L76
 
 | | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Jhan, C.R, Satange, R.B, Lin, S.M. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
4CPZ
 
 | | Structure of the Neuraminidase from the B/Lyon/CHU/15.216/2011 virus in complex with Zanamivir | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | Deposit date: | 2014-02-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
4CPL
 
 | | Structure of the Neuraminidase from the B/Brisbane/60/2008 virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | Deposit date: | 2014-02-07 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
4CPO
 
 | | Structure of the Neuraminidase from the B/Lyon/CHU/15.216/2011 virus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | Deposit date: | 2014-02-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
