8S8J
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF5) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8RW1
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8E
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8F
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
5BR7
 
 | | Structure of UDP-galactopyranose mutase from Corynebacterium diphtheriae in complex with citrate ion | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wangkanont, K, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational Control of UDP-Galactopyranose Mutase Inhibition.
Biochemistry, 56, 2017
|
|
5LFO
 
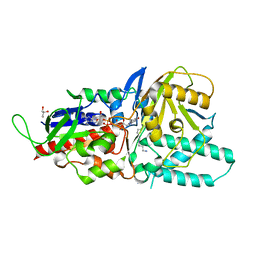 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LGB
 
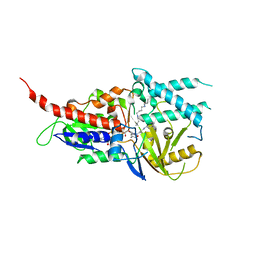 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with MDL72527 | | Descriptor: | FAD-MDL72527 adduct, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-06 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5MBX
 
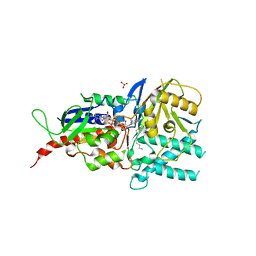 | | Crystal structure of reduced murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
3KYB
 
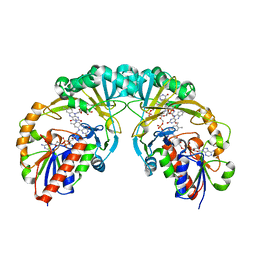 | |
8K8T
 
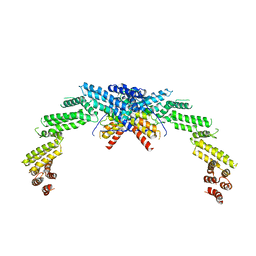 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8OGD
 
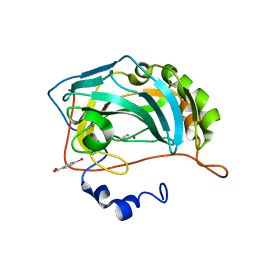 | | Structure of zinc(II) double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, THIOCYANATE ION, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
8OGE
 
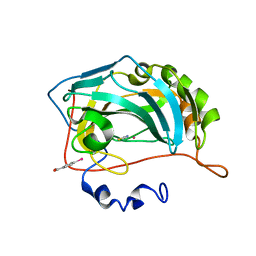 | | Structure of cobalt(II) substituted double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
3MMJ
 
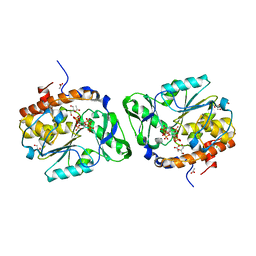 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol hexakisphosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-04-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding in protein-tyrosine phosphatase-like inositol polyphosphatases.
J.Biol.Chem., 287, 2012
|
|
6YPO
 
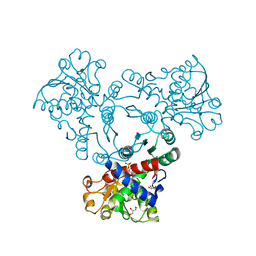 | | Arabidopsis aspartate transcarbamoylase bound to UMP | | Descriptor: | GLYCEROL, PYRB, URIDINE-5'-MONOPHOSPHATE | | Authors: | Ramon Maiques, S, Del Cano Ochoa, F, Bellin, L, Mohlmann, T. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase.
Nat Commun, 12, 2021
|
|
5D8N
 
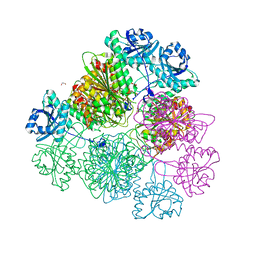 | | Tomato leucine aminopeptidase mutant - K354E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | DuPrez, K.T, Scranton, M.A, Walling, L.L, Fan, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into chaperone-activity enhancement by a K354E mutation in tomato acidic leucine aminopeptidase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6YEL
 
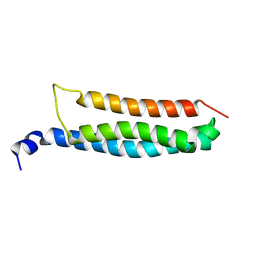 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
8PHM
 
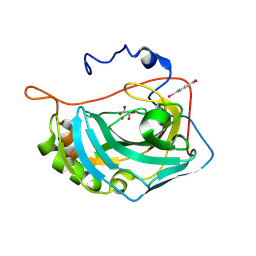 | | Oxalate-bound cobalt(II) human carbonic anhydrase II | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Gigli, L, Malanho Silva, J, Cerofolini, L, Macedo, A.L, Geraldes, C.F.G.C, Suturina, E.A, Calderone, V, Fragai, M, Parigi, G, Ravera, E, Luchinat, C. | | Deposit date: | 2023-06-20 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Machine Learning-Enhanced Quantum Chemistry-Assisted Refinement of the Active Site Structure of Metalloproteins.
Inorg.Chem., 63, 2024
|
|
8G4A
 
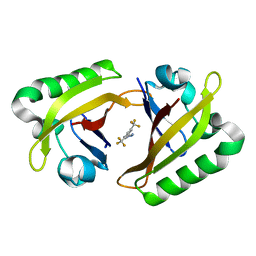 | |
3MOZ
 
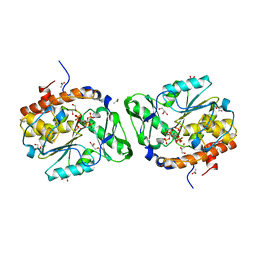 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol (1,2,3,5,6)pentakisphosphate | | Descriptor: | (1R,2R,3R,4R,5S,6S)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], ACETATE ION, CHLORIDE ION, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-04-23 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding in protein-tyrosine phosphatase-like inositol polyphosphatases.
J.Biol.Chem., 287, 2012
|
|
3KHL
 
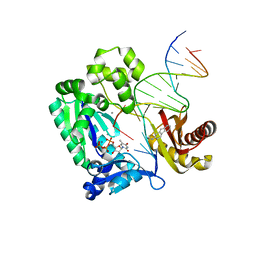 | | Dpo4 post-extension ternary complex with misinserted A opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2-AMINOFLUORENE, 5'-D(*CP*C*TP*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*AP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KHR
 
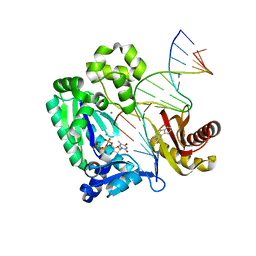 | | Dpo4 post-extension ternary complex with the correct C opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2-AMINOFLUORENE, 5'-D(*CP*C*TP*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*CP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5CZL
 
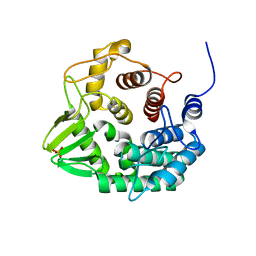 | | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library | | Descriptor: | Glucanase, PHOSPHATE ION | | Authors: | Scapin, S.M.N, Souza, F.H.M, Zanphorlin, L.M, Almeida, T.S, Sade, Y.B, Cardoso, A.M, Pinheiro, G.L, Murakami, M.T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library
To Be Published
|
|
6HQU
 
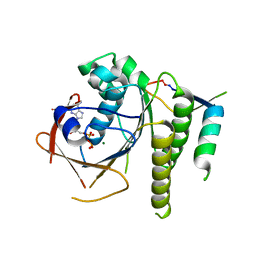 | | Humanised RadA mutant HumRadA22 in complex with a recombined BRC repeat 8-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Breast cancer type 2 susceptibility, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Lindenburg, L, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Improved RAD51 binders through motif shuffling based on the modularity of BRC repeats.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8PU1
 
 | |
