6XR2
 
 | |
6XR1
 
 | |
4YIT
 
 | |
4YIS
 
 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | 著者 | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | 登録日 | 2015-03-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4Z1Z
 
 | |
4Z20
 
 | |
4YHX
 
 | |
5ESP
 
 | |
4YY5
 
 | |
4YY2
 
 | |
5THG
 
 | |
5KW2
 
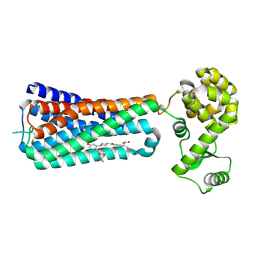 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | 分子名称: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | 著者 | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | 登録日 | 2016-07-15 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
3WTB
 
 | |
3WTC
 
 | |
1D8B
 
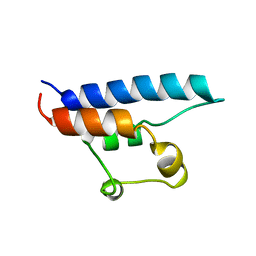 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | 分子名称: | SGS1 RECQ HELICASE | | 著者 | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | 登録日 | 1999-10-21 | | 公開日 | 2000-01-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
1DZ7
 
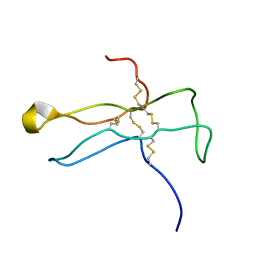 | | Solution structure of the a-subunit of human chorionic gonadotropin [modeled without carbohydrate residues] | | 分子名称: | CHORIONIC GONADOTROPIN | | 著者 | Erbel, P.J.A, Karimi-Nejad, Y, De Beer, T, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | 登録日 | 2000-02-18 | | 公開日 | 2000-02-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the alpha-subunit of human chorionic gonadotropin.
Eur.J.Biochem., 260, 1999
|
|
6PNR
 
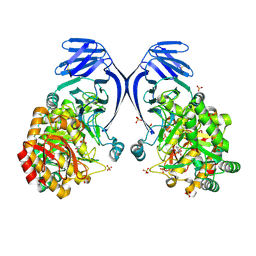 | | A GH31 family sulfoquinovosidase from E. rectale in complex with aza-sugar inhibitor IFGSQ | | 分子名称: | Alpha-glucosidase, SULFATE ION, [(3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)piperidin-3-yl]methanesulfonic acid | | 著者 | Jarva, M.A, Lingford, J.P, John, A, Goddard-Borger, E.D. | | 登録日 | 2019-07-03 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A GH31 family sulfoquinovosidase from E. rectale in complex with aza-sugar inhibitor IFGSQ
To Be Published
|
|
4WHA
 
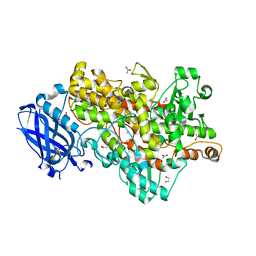 | | Lipoxygenase-1 (soybean) L546A/L754A mutant | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, FE (III) ION, ... | | 著者 | Scouras, A.D, Carr, C.A.M, Hu, S, Klinman, J.P. | | 登録日 | 2014-09-21 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Extremely elevated room-temperature kinetic isotope effects quantify the critical role of barrier width in enzymatic C-H activation.
J.Am.Chem.Soc., 136, 2014
|
|
4WFO
 
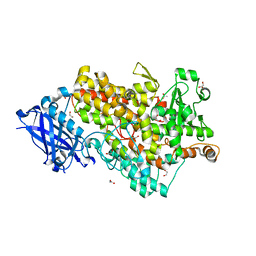 | |
4WSL
 
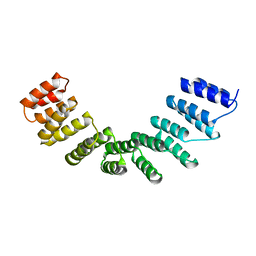 | | Crystal structure of designed cPPR-polyC protein | | 分子名称: | Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-10-28 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4WN4
 
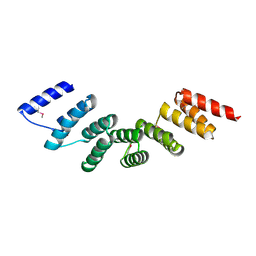 | | Crystal structure of designed cPPR-polyA protein | | 分子名称: | Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-10-10 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJS
 
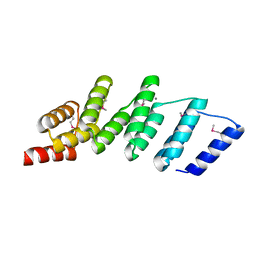 | | Crystal structure of designed (SeMet)-cPPR-NRE protein | | 分子名称: | CALCIUM ION, Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-05-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJQ
 
 | | Crystal structure of designed cPPR-polyG protein | | 分子名称: | Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-05-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.353 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJR
 
 | | Crystal structure of designed cPPR-NRE protein | | 分子名称: | MAGNESIUM ION, Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-05-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
3N9H
 
 | | Crystal Structural of mutant Y305A in the copper amine oxidase from hansenula polymorpha | | 分子名称: | COPPER (II) ION, Peroxisomal primary amine oxidase | | 著者 | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | 登録日 | 2010-05-30 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
