7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6KLY
 
 | | Crystal structure of the type III effector XopAI from Xanthomonas axonopodis pv. citri in space group P43212 | | Descriptor: | Type III effector XopAI | | Authors: | Liu, J.-H, Wu, J.E, Lin, H, Chiu, S.W, Yang, J.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure-Based Exploration of Arginine-Containing Peptide Binding in the ADP-Ribosyltransferase Domain of the Type III Effector XopAI Protein.
Int J Mol Sci, 20, 2019
|
|
6BXM
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM/cleaved SAM | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALPHA-AMINOBUTYRIC ACID, Diphthamide biosynthesis enzyme Dph2, ... | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXN
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXO
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAH | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXL
 
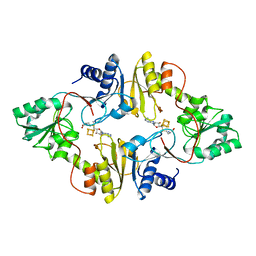 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
3I9N
 
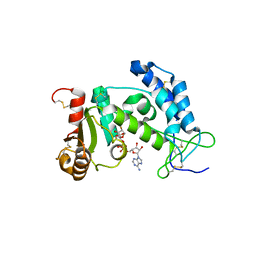 | | Crystal structure of human CD38 complexed with an analog ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9K
 
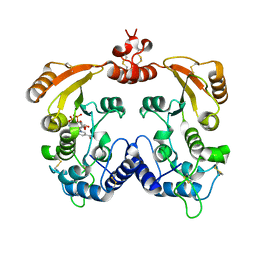 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
6BXK
 
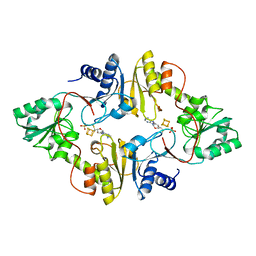 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and MTA | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
3I9J
 
 | | Crystal structure of ADP ribosyl cyclase complexed with a substrate analog and a product nicotinamide | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE, Nicotinamide 2-fluoro-adenine dinucleotide, ... | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9L
 
 | | Crystal structure of ADP ribosyl cyclase complexed with N1-cIDPR | | Descriptor: | ADP-ribosyl cyclase, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9M
 
 | | Crystal structure of human CD38 complexed with an analog ara-2'F-ADPR | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3U57
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5Y
 
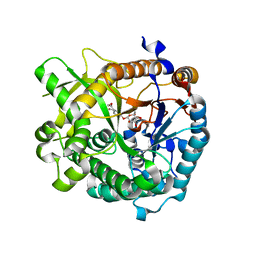 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
7XKE
 
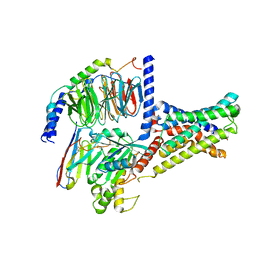 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
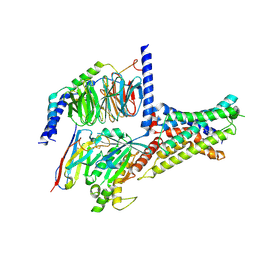 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
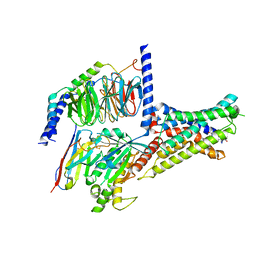 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
3U5U
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
8TMT
 
 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TN0
 
 | | Crystal structure of KPC-44 carbapenemase w/o cryoprotectant | | Descriptor: | SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJM
 
 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TMR
 
 | | Crystal structure of KPC-44 carbapenemase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
3DZH
 
 | | Crystal structure of human CD38 extracellular domain, GTP complex | | Descriptor: | ADP-ribosyl cyclase 1, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Liu, Q, Kriksunov, I.A, Jiang, H, Graeff, R, Lin, H, Lee, H.C, Hao, Q. | | Deposit date: | 2008-07-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Covalent and Noncovalent Intermediates of an NAD Utilizing Enzyme, Human CD38.
Chem.Biol., 15, 2008
|
|
3DZJ
 
 | | Crystal structure of human CD38 extracellular domain E226Q mutant, NMN complex | | Descriptor: | ADP-ribosyl cyclase 1, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE | | Authors: | Liu, Q, Kriksunov, I.A, Jiang, H, Graeff, R, Lin, H, Lee, H.C, Hao, Q. | | Deposit date: | 2008-07-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Covalent and Noncovalent Intermediates of an NAD Utilizing Enzyme, Human CD38.
Chem.Biol., 15, 2008
|
|
