7KPF
 
 | | NME2 bound to myristoyl-CoA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate kinase B, ... | | Authors: | Price, I.R, Lin, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical Proteomic Approach reveals the regulation of NME1/2 and cancer metastasis by Long-Chain Fatty Acyl Coenzyme A
To Be Published
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7X8H
 
 | | Crystal structure of AtHPPD-(+)-Usnic acid complex | | Descriptor: | (9bR)-2,6-diethanoyl-8,9b-dimethyl-3,7,9-tris(oxidanyl)dibenzofuran-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2022-03-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X8D
 
 | | Crystal structure of AtHPPD-phenylpyruvate complex | | Descriptor: | 3-PHENYLPYRUVIC ACID, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2022-03-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X8I
 
 | | Crystal structure of AtHPPD-Shikonin complex | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2022-03-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7MDO
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
7MDM
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
7WJ8
 
 | | Complex structure of AtHPPD-PyQ1 | | Descriptor: | 2-pyren-1-yloxyethyl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-01-05 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WJJ
 
 | | Complex structure of AtHPPD-PyQ2 | | Descriptor: | 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]-N-(2-pyren-1-yloxyethyl)ethanamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-01-06 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7VC8
 
 | | Complex structure of AtHPPD with inhibitor PYQ3 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, pyren-1-yl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate | | Authors: | Yang, G.F, Lin, H.Y. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WJK
 
 | | Complex structure of AtHPPD-ZWJ | | Descriptor: | 3-[2-[4-[[4-[2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9,11-tetraen-8-yl]phenoxy]methyl]-1,2,3-triazol-1-yl]ethyl]-1,5-dimethyl-6-[(1S)-2-oxidanyl-6-oxidanylidene-cyclohex-2-en-1-yl]carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-01-06 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Complex structure of AtHPPD-PyQ1
To Be Published
|
|
6ZHU
 
 | | Yeast Uba1 in complex with Ubc3 and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system
To Be Published
|
|
6YK0
 
 | |
6YJZ
 
 | |
6YK1
 
 | |
6ZJC
 
 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione and triethyltin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6ZHS
 
 | | Uba1 bound to two E2 (Ubc13) molecules | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHT
 
 | | Uba1-Ubc13 disulfide mediated complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Schaefer, A, Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
8FXT
 
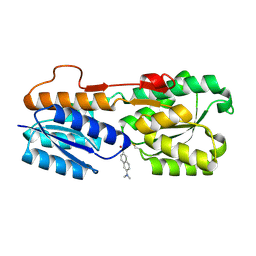 | | Escherichia coli periplasmic Glucose-Binding Protein glucose complex: Acrylodan conjugate attached at W183C | | Descriptor: | 1-[6-(dimethylamino)naphthalen-2-yl]propan-1-one, CALCIUM ION, D-galactose/methyl-galactoside binding periplasmic protein MglB, ... | | Authors: | Allert, M.J, Kumar, S, Wang, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Chromophore carbonyl twisting in fluorescent biosensors encodes direct readout of protein conformations with multicolor switching.
Commun Chem, 6, 2023
|
|
8FXU
 
 | | Thermoanaerobacter thermosaccharolyticum periplasmic Glucose-Binding Protein glucose complex: Badan conjugate attached at F17C | | Descriptor: | 2-bromo-1-[6-(dimethylamino)naphthalen-2-yl]ethan-1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Allert, M.J, Kumar, S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Chromophore carbonyl twisting in fluorescent biosensors encodes direct readout of protein conformations with multicolor switching.
Commun Chem, 6, 2023
|
|
6ZJ9
 
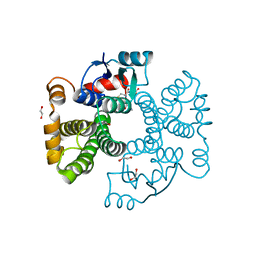 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6ZQH
 
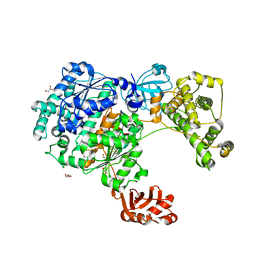 | | Yeast Uba1 in complex with ubiquitin | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Chemistry, 27, 2021
|
|
7ZH9
 
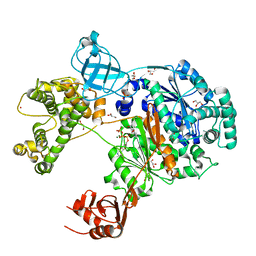 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
