4Q7B
 
 | |
1YTQ
 
 | |
2BKY
 
 | | Crystal structure of the Alba1:Alba2 heterodimer from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA/RNA-BINDING PROTEIN ALBA 1, DNA/RNA-BINDING PROTEIN ALBA 2 | | Authors: | Jelinska, C, Conroy, M.J, Craven, C.J, Bullough, P.A, Waltho, J.P, Taylor, G.L, White, M.F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Obligate Heterodimerization of the Archaeal Alba2 Protein with Alba1 Provides a Mechanism for Control of DNA Packaging.
Structure, 13, 2005
|
|
4D0A
 
 | |
8Y2U
 
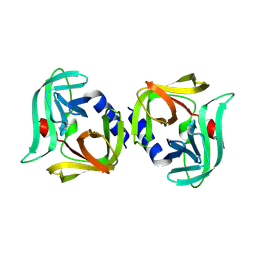 | | Crystal structure of 3C protease from coxsackievirus B4 | | Descriptor: | Protease 3C | | Authors: | Jiang, H.H, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of the 3C proteases from Coxsackievirus B3 and B4.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
7NNU
 
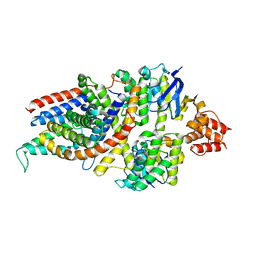 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs | | Descriptor: | Conserved hypothetical membrane protein, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NNT
 
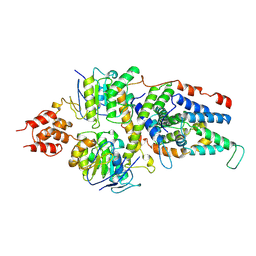 | | Cryo-EM structure of the folate-specific ECF transporter complex in DDM micelles | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8CXC
 
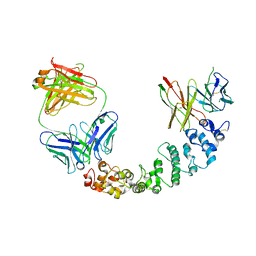 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
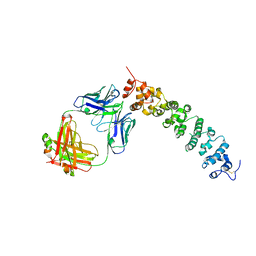 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
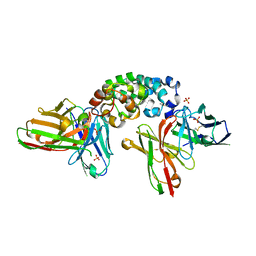 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8DD7
 
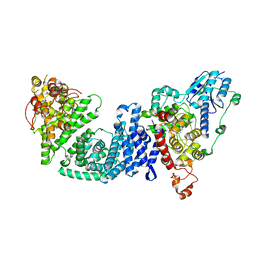 | | The Cryo-EM structure of Drosophila Cryptochrome in complex with Timeless | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylated-DNA--protein-cysteine methyltransferase,Cryptochrome-1 fusion, Protein timeless,Methylated-DNA--protein-cysteine methyltransferase fusion | | Authors: | Feng, S, Lin, C, DeOliveira, C.C, Crane, B.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryptochrome-Timeless structure reveals circadian clock timing mechanisms.
Nature, 617, 2023
|
|
6JJ0
 
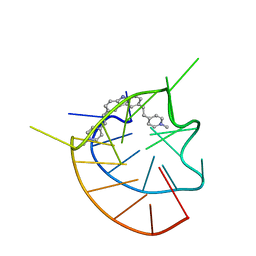 | |
4V9Q
 
 | | Crystal Structure of Blasticidin S Bound to Thermus Thermophilus 70S Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Ling, C, Ermolenko, D.N, Korostelev, A.A. | | Deposit date: | 2013-06-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3NW2
 
 | | Novel nanomolar Imidazopyridines as selective Nitric Oxide Synthase (iNOS) inhibitors: SAR and structural insights | | Descriptor: | 2-[2-(4-methoxypyridin-2-yl)ethyl]-3H-imidazo[4,5-b]pyridine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Graedler, U, Fuchss, T, Ulrich, W.R, Boer, R, Strub, A, Hesslinger, C, Anezo, C, Diederichs, K, Zaliani, A. | | Deposit date: | 2010-07-09 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel nanomolar imidazo[4,5-b]pyridines as selective nitric oxide synthase (iNOS) inhibitors: SAR and structural insights
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6ECZ
 
 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | Carbonic anhydrase 2, N-[2-hydroxy-3-nitro-5-(nitrosulfonyl)phenyl]-N'-(pentafluorophenyl)urea, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
8J3A
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
Y54C mutant in complex with PF00835231
To Be Published
|
|
8J37
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF00835231
To Be Published
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF00835231
To Be Published
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231
To Be Published
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF00835231
To Be Published
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF00835231
To Be Published
|
|
8J39
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF00835231
To Be Published
|
|
8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | Descriptor: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of MERS main protease in complex with PF00835231
To Be Published
|
|
8J36
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF00835231
To Be Published
|
|
4LX2
 
 | | Crystal structure of Myo5a globular tail domain in complex with melanophilin GTBD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Melanophilin, Unconventional myosin-Va | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Houdusse, A. | | Deposit date: | 2013-07-29 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
