4QX4
 
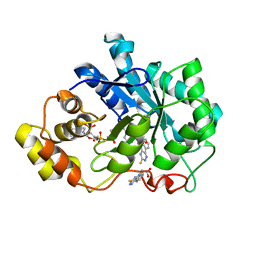 | | Human Aldose Reductase complexed with a ligand with a new scaffold at 1.26 A | | Descriptor: | (3-thioxo-2,3-dihydro-5H-[1,2,4]triazino[5,6-b]indol-5-yl)acetic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2014-07-18 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.259 Å) | | Cite: | Identification of novel aldose reductase inhibitors based on carboxymethylated mercaptotriazinoindole scaffold.
J.Med.Chem., 58, 2015
|
|
4QR6
 
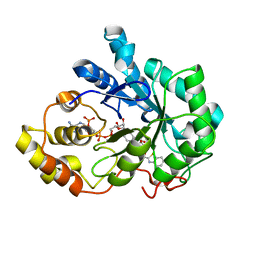 | |
4ES9
 
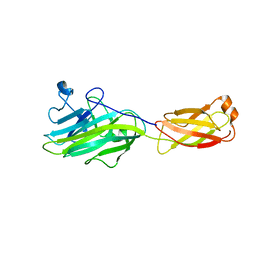 | |
6MBY
 
 | | Carbonic Anhydrase II in complex with Ferulic Acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, McKenna, R. | | Deposit date: | 2018-08-30 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Carbonic anhydrase II in complex with carboxylic acid-based inhibitors.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6MBV
 
 | |
6B4D
 
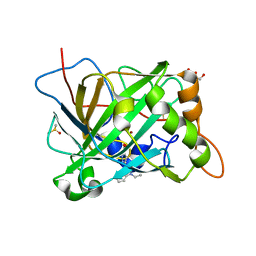 | | Crystal structure of human carbonic anhydrase II in complex with a heteroaryl-pyrazole carboxylic acid derivative. | | Descriptor: | 3-(1-ethyl-1H-indol-3-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, R. | | Deposit date: | 2017-09-26 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.196 Å) | | Cite: | Exploring Heteroaryl-pyrazole Carboxylic Acids as Human Carbonic Anhydrase XII Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6B5A
 
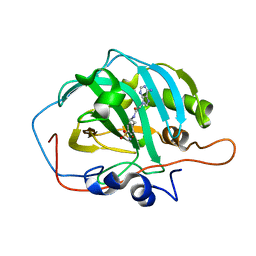 | |
6B59
 
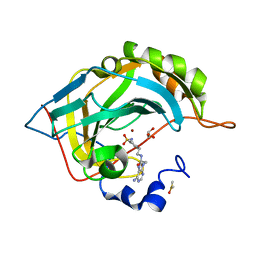 | |
7OXX
 
 | | CrabP2 mutant R30AK31A | | Descriptor: | Cellular retinoic acid-binding protein 2, SODIUM ION | | Authors: | Tomlinson, C.W.E, Basle, A, Pohl, E. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural requirements for the specific binding of CRABP2 to cyclin D3
To Be Published
|
|
2BKY
 
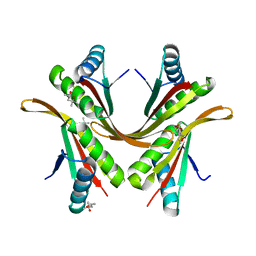 | | Crystal structure of the Alba1:Alba2 heterodimer from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA/RNA-BINDING PROTEIN ALBA 1, DNA/RNA-BINDING PROTEIN ALBA 2 | | Authors: | Jelinska, C, Conroy, M.J, Craven, C.J, Bullough, P.A, Waltho, J.P, Taylor, G.L, White, M.F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Obligate Heterodimerization of the Archaeal Alba2 Protein with Alba1 Provides a Mechanism for Control of DNA Packaging.
Structure, 13, 2005
|
|
7NGG
 
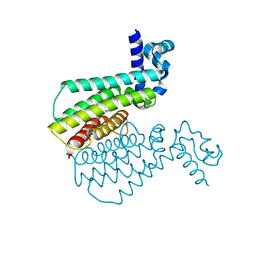 | |
6HKR
 
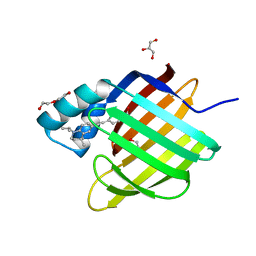 | | Human Cellular Retinoic Acid Binding Protein II (CRABPII) with bound synthetic retinoid DC271. | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4,4-dimethyl-1-propan-2-yl-2,3-dihydroquinolin-6-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2, ... | | Authors: | Tomlinson, C, Chisholm, D, Whiting, A, Pohl, E. | | Deposit date: | 2018-09-07 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Fluorescence Competition Assay for Retinoic Acid Binding Proteins.
ACS Med Chem Lett, 9, 2018
|
|
6CJV
 
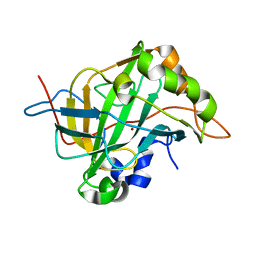 | | Carbonic anhydrase IX-mimic in complex with sucralose | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Carbonic anhydrase 2, ZINC ION | | Authors: | Lomelino, C.L, Murray, A.B, McKenna, R. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Sweet Binders: Carbonic Anhydrase IX in Complex with Sucralose.
ACS Med Chem Lett, 9, 2018
|
|
6O2W
 
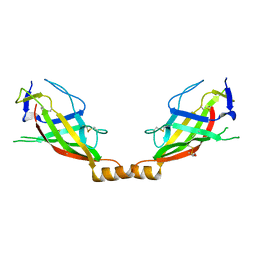 | |
6O2U
 
 | | Crystal structure of the SARAF luminal domain | | Descriptor: | Store-operated calcium entry-associated regulatory factor | | Authors: | Kimberlin, C.R, Minor, D.L. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARAF Luminal Domain Structure Reveals a Novel Domain-Swapped beta-Sandwich Fold Important for SOCE Modulation.
J.Mol.Biol., 431, 2019
|
|
6O2V
 
 | |
3GI1
 
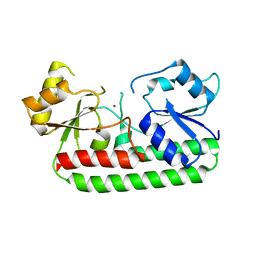 | | Crystal Structure of the laminin-binding protein Lbp of Streptococcus pyogenes | | Descriptor: | Laminin-binding protein of group A streptococci, ZINC ION | | Authors: | Linke, C, Caradoc-Davies, T.T, Young, P.G, Proft, T, Baker, E.N. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The laminin-binding protein Lbp from Streptococcus pyogenes is a zinc receptor
J.Bacteriol., 191, 2009
|
|
4ES8
 
 | | Crystal Structure of the adhesin domain of Epf from Streptococcus pyogenes in P212121 | | Descriptor: | ACETATE ION, Epf, GLYCEROL, ... | | Authors: | Linke, C, Siemens, N, Kreikemeyer, B, Baker, E.N. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Extracellular Protein Factor Epf from Streptococcus pyogenes Is a Cell Surface Adhesin That Binds to Cells through an N-terminal Domain Containing a Carbohydrate-binding Module.
J.Biol.Chem., 287, 2012
|
|
3KS8
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
1EOK
 
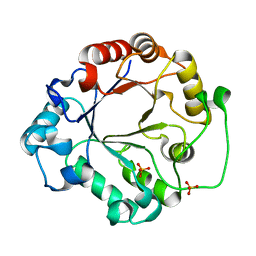 | | CRYSTAL STRUCTURE OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3 | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3, SULFATE ION | | Authors: | Waddling, C.A, Plummer Jr, T.H, Tarentino, A.L, Van Roey, P. | | Deposit date: | 2000-03-23 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3).
Biochemistry, 39, 2000
|
|
3EH0
 
 | | Crystal Structure of LpxD from Escherichia coli | | Descriptor: | UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Bartling, C.M, Raetz, C.R.H. | | Deposit date: | 2008-09-11 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis
Biochemistry, 48, 2009
|
|
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
3KLQ
 
 | | Crystal Structure of the Minor Pilin FctB from Streptococcus pyogenes 90/306S | | Descriptor: | GLYCEROL, Putative pilus anchoring protein | | Authors: | Linke, C, Young, P.G, Bunker, R.D, Caradoc-Davies, T.T, Baker, E.N. | | Deposit date: | 2009-11-08 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring
J.Biol.Chem., 285, 2010
|
|
1EOM
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3 WITH A BIANTENNARY COMPLEX OCTASACCHARIDE | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3, SULFATE ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Waddling, C.A, Plummer Jr, T.H, Tarentino, A.L, Van Roey, P. | | Deposit date: | 2000-03-23 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3).
Biochemistry, 39, 2000
|
|
5WLU
 
 | |
