6LDD
 
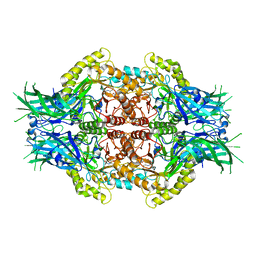 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-propyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEJ
 
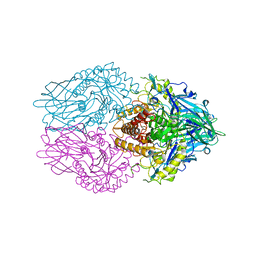 | | Structure of E. coli beta-glucuronidase complex with C6-propyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.617 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEL
 
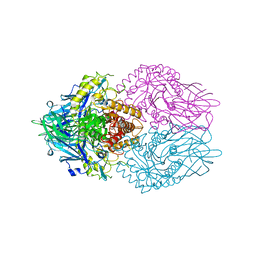 | | Structure of E. coli beta-glucuronidase complex with C6-hexyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-hexyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LDC
 
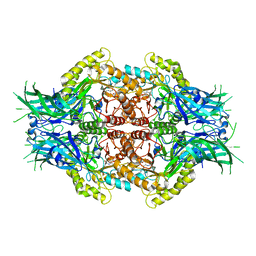 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-nonyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-nonyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
3KCW
 
 | | Crystal structure of Ganoderma fungal immunomodulatory protein, GMI | | Descriptor: | immunomodulatory protein | | Authors: | Hsu, M.F, Wang, A.H.J, Yang, C.S, Huang, C.T, Hseu, R.S, Lin, C.W, Wu, M.Y, Huang, C.S, Fu, H.Y. | | Deposit date: | 2009-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single cysteine replacement at Leu6 increase the potent and thermostability in Ganoderma fungal immunomodulatory proteins
To be Published
|
|
1HWV
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
9C8X
 
 | |
9C90
 
 | |
9C8Z
 
 | |
9C8Y
 
 | |
9C8W
 
 | |
8CXC
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
7XGZ
 
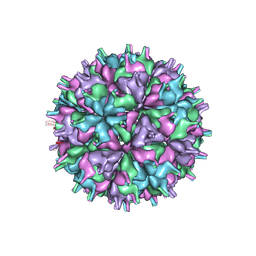 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
8J3A
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
Y54C mutant in complex with PF00835231
To Be Published
|
|
8J37
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF00835231
To Be Published
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF00835231
To Be Published
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231
To Be Published
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF00835231
To Be Published
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF00835231
To Be Published
|
|
8J39
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF00835231
To Be Published
|
|
8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | Descriptor: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of MERS main protease in complex with PF00835231
To Be Published
|
|
8J36
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF00835231
To Be Published
|
|
