6POR
 
 | | Antimicrobial lasso peptide ubonodin | | Descriptor: | Ubonodin | | Authors: | Link, A.J, Cheung-Lee, W.L. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Discovery of Ubonodin, an Antimicrobial Lasso Peptide Active against Members of the Burkholderia cepacia Complex.
Chembiochem, 21, 2020
|
|
2N68
 
 | |
8DYM
 
 | | Aspartimidylated Graspetide Amycolimiditide | | Descriptor: | ATP-grasp target RiPP | | Authors: | Link, A.J, Choi, B. | | Deposit date: | 2022-08-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanistic Analysis of the Biosynthesis of the Aspartimidylated Graspetide Amycolimiditide.
J.Am.Chem.Soc., 144, 2022
|
|
6B5W
 
 | | Benenodin-1-dC5, state 2 | | Descriptor: | Benenodin-1 | | Authors: | Link, A.J, Zong, C. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Lasso Peptide Benenodin-1 Is a Thermally Actuated [1]Rotaxane Switch
J. Am. Chem. Soc., 139 (30), 2017
|
|
6Q1X
 
 | | Lasso peptide pandonodin | | Descriptor: | Pandonodin | | Authors: | Link, A.J, Cheung-Lee, W.L. | | Deposit date: | 2019-08-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pandonodin: A Proteobacterial Lasso Peptide with an Exceptionally Long C-Terminal Tail.
Acs Chem.Biol., 14, 2019
|
|
5KTC
 
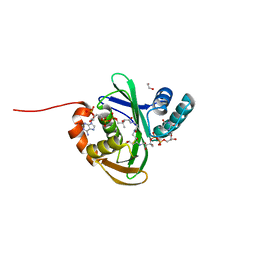 | | FdhC with bound products: Coenzyme A and 3-[(R)-3-hydroxybutanoylamino]-3,6-dideoxy-d-galactose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
5KTA
 
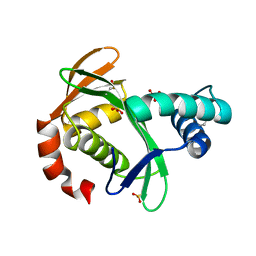 | | Apo FdhC- a nucleotide-linked sugar GNAT | | Descriptor: | DIMETHYL SULFOXIDE, FdhC, SULFATE ION | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
5KTD
 
 | | FdhC with bound products: Coenzyme A and dTDP-3-amino-3,6-dideoxy-d-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
6MW6
 
 | |
7LCW
 
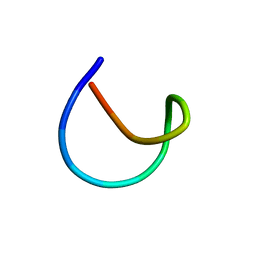 | | Aspartimidylated Lasso Peptide Lihuanodin | | Descriptor: | Lasso Peptide Lihuanodin | | Authors: | Link, A.J, Cao, L. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cellulonodin-2 and Lihuanodin: Lasso Peptides with an Aspartimide Post-Translational Modification.
J.Am.Chem.Soc., 143, 2021
|
|
7LL7
 
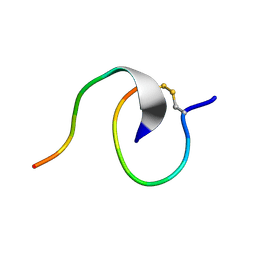 | | [2]Catenane From MccJ25 Variant G12C G21C | | Descriptor: | GLY-GLY-ALA-GLY-HIS-VAL-PRO-GLU-TYR-PHE, VAL-CYS-ILE-GLY-THR-PRO-ILE-SER-PHE-TYR-CYS | | Authors: | Link, A.J, Schroeder, H.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Dynamic covalent self-assembly of mechanically interlocked molecules solely made from peptides.
Nat.Chem., 13, 2021
|
|
7LIF
 
 | |
7LI2
 
 | | Omega ester peptide pre-fuscimiditide | | Descriptor: | Pre-fuscimiditide peptide | | Authors: | Link, A.J, Elashal, H.E. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis and characterization of fuscimiditide, an aspartimidylated graspetide.
Nat.Chem., 14, 2022
|
|
8VYC
 
 | |
5T56
 
 | |
7U4L
 
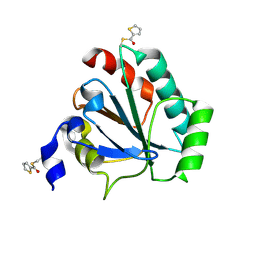 | | Crystal structure of human GPX4-U46C in complex with MAC-5576 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, thiophene-2-carbaldehyde | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4K
 
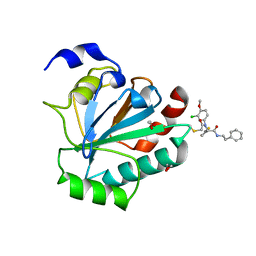 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4N
 
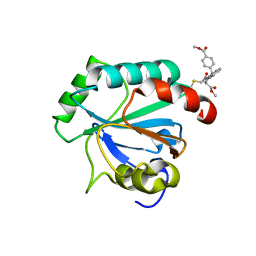 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4M
 
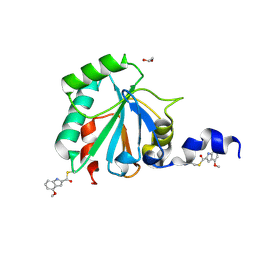 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4J
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with TMT10 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4I
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | Descriptor: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
6D0H
 
 | |
6D0I
 
 | | ParT: Prs ADP-ribosylating toxin bound to cognate antitoxin ParS. L48M ParT, SeMet-substituted complex. | | Descriptor: | GLYCEROL, ParS: COG5642 (DUF2384) antitoxin fragment, ParT: COG5654 (RES domain) toxin | | Authors: | Piscotta, F.J, Jeffrey, P.D, Link, A.J. | | Deposit date: | 2018-04-10 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ParST is a widespread toxin-antitoxin module that targets nucleotide metabolism.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8DYN
 
 | |
8SVB
 
 | | Antimicrobial lasso peptide achromonodin-1 | | Descriptor: | Achromonodin-1 | | Authors: | Carson, D.V, Cheung-Lee, W.L, So, L, Link, A.J. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Bioactivity of the Achromonodins: Lasso Peptides Encoded by Achromobacter .
J.Nat.Prod., 86, 2023
|
|
