5EPP
 
 | | Structural Insights into the Interaction of p97 N-terminus Domain and VBM Motif in Rhomboid Protease, RHBDL4 | | Descriptor: | Rhomboid-related protein 4, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Ly, T.T, Kang, J.Y, Lee, J.-G, An, J.Y, Youn, H.-S, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2015-11-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into the interaction of p97 N-terminus domain and VBM in rhomboid protease, RHBDL4.
Biochem.J., 473, 2016
|
|
8TRL
 
 | | T cell recognition of citrullinated alpha-enolase peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
3RVG
 
 | | Crystals structure of Jak2 with a 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitor | | Descriptor: | 1-(cyclohexylamino)-7-(1-methyl-1H-pyrazol-4-yl)-5H-pyrido[4,3-b]indole-4-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Lim, J, Taoka, B, Otte, R.D, Spencer, K, Dinsmore, C.J, Altman, M.D, Chan, G, Rosenstein, C, Sharma, S, Su, H.P, Szewczak, A.A, Xu, L, Yin, H, Zugay-Murphy, J, Marshall, C.G, Young, J.R. | | Deposit date: | 2011-05-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Discovery of 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitors of Janus kinase 2 (JAK2) for the treatment of myeloproliferative disorders.
J.Med.Chem., 54, 2011
|
|
5GLF
 
 | | Structural insights into the interaction of p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease | | Descriptor: | Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Yoon, S.Y, Ly, T.T, Kang, J.Y, Youn, H.-S, An, J.Y, Lee, J.-G, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2016-07-11 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease.
FEBS Lett., 590, 2016
|
|
6V1A
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 74cit69-81, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
6V13
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 74cit69-81, G08 TCR alpha chain, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
6V0Y
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 72,74cit69-81, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
6V15
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 72,74cit69-81, G08 TCR alpha chain, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
6V19
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 72,74cit69-81, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
6V18
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
7RDV
 
 | |
7T6I
 
 | | Crystal structure of HLA-DP1 in complex with pp65 peptide in reverse orientation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Lim, J.J, Reid, H, Rossjohn, J. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human T cells recognize HLA-DP-bound peptides in two orientations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2L9F
 
 | | NMR solution structure of meACP | | Descriptor: | CalE8 | | Authors: | Lim, J, Yang, D, Liang, Z.X, Kong, R, Murugan, E, Ho, C.L. | | Deposit date: | 2011-02-08 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the acyl carrier protein domain from the highly reducing type I iterative polyketide synthase CalE8
Plos One, 6, 2011
|
|
2LUT
 
 | | NMR solution structure of midkine-a | | Descriptor: | Midkine-related growth factor | | Authors: | Lim, J, Yang, D, Meng, D. | | Deposit date: | 2012-06-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Two Zebrafish Midkine Proteins Reveals Importance of the Conserved Hinge for Heparin Binding and Embryogenesis
To be Published
|
|
2M6M
 
 | |
2CB3
 
 | | Crystal structure of peptidoglycan recognition protein-LE in complex with tracheal cytotoxin (monomeric diaminopimelic acid-type peptidoglycan) | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, GLYCEROL, PEPTIDOGLYCAN-RECOGNITION PROTEIN-LE | | Authors: | Lim, J.-H, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2005-12-29 | | Release date: | 2006-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Preferential Recognition of Diaminopimelic Acid-Type Peptidoglycan by a Subset of Peptidoglycan Recognition Proteins
J.Biol.Chem., 281, 2006
|
|
1COJ
 
 | | FE-SOD FROM AQUIFEX PYROPHILUS, A HYPERTHERMOPHILIC BACTERIUM | | Descriptor: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | Authors: | Lim, J.H, Yu, Y.G, Kim, S.-H, Cho, S.-J, Ahn, B.Y, Han, Y.S, Cho, Y. | | Deposit date: | 1999-05-28 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability.
J.Mol.Biol., 270, 1997
|
|
5BZ6
 
 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4MM2
 
 | | Crystal structure of yeast primase catalytic subunit | | Descriptor: | CADMIUM ION, CITRIC ACID, DNA primase small subunit | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-09-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast primase catalytic subunit
To be Published
|
|
2QJ6
 
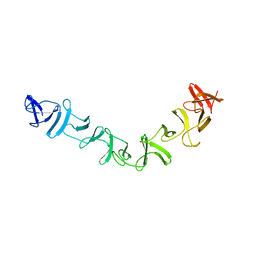 | | Crystal structure analysis of a 14 repeat C-terminal fragment of toxin TcdA in Clostridium difficile | | Descriptor: | Toxin A | | Authors: | Albesa-Jove, D, Bertrand, T, Carpenter, L, Lim, J, Brown, K.A, Fairweather, N. | | Deposit date: | 2007-07-06 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solution and crystal structures of the cell binding domain of toxins TcdA and TcdB from Clostridium difficile
To be Published
|
|
4XSJ
 
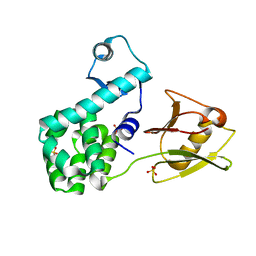 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
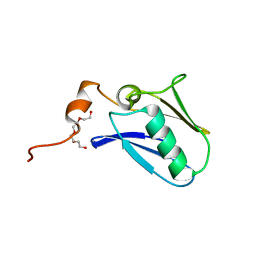 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
5KE0
 
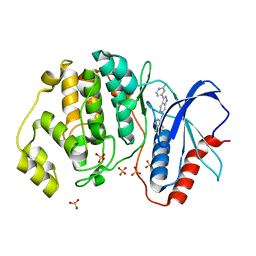 | | Discovery of 1-1H-Pyrazolo 4,3-c pyridine-6-yl urea Inhibitors of Extracellular Signal Regulated Kinase ERK for the Treatment of Cancers | | Descriptor: | 1-[3-(2-methylpyridin-4-yl)-1~{H}-pyrazolo[4,3-c]pyridin-6-yl]-3-(phenylmethyl)urea, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Lim, J. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of 1-(1H-Pyrazolo[4,3-c]pyridin-6-yl)urea Inhibitors of Extracellular Signal-Regulated Kinase (ERK) for the Treatment of Cancers.
J.Med.Chem., 59, 2016
|
|
4WYS
 
 | | Crystal structure of thiolase from Escherichia coli | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4WYR
 
 | | Crystal structure of thiolase mutation (V77Q,N153Y,A286K) from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
