3NG2
 
 | |
2XFL
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Sun, H, Bricogne, G, Liang, Z, Lescar, J. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
2XEM
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | (3E,5E,7E,9E,11E,13E)-pentadeca-3,5,7,9,11,13-hexaen-2-one, DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Bricogne, G, Liang, Z.X, Lescar, J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
1FV5
 
 | | SOLUTION STRUCTURE OF THE FIRST ZINC FINGER FROM THE DROSOPHILA U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | FIRST ZINC FINGER OF U-SHAPED, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1FU9
 
 | | SOLUTION STRUCTURE OF THE NINTH ZINC-FINGER DOMAIN OF THE U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | U-SHAPED TRANSCRIPTIONAL COFACTOR, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
2JM3
 
 | | Solution structure of the THAP domain from C. elegans C-terminal binding protein (CtBP) | | Descriptor: | Hypothetical protein, ZINC ION | | Authors: | Liew, C.K, Crossley, M, Mackay, J.P, Nicholas, H.R. | | Deposit date: | 2006-09-21 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the THAP domain from Caenorhabditis elegans C-terminal binding protein (CtBP).
J.Mol.Biol., 366, 2007
|
|
4AMO
 
 | |
4AMN
 
 | |
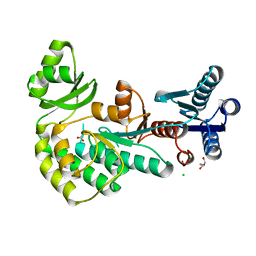
4AMP
 
 | |
4AMM
 
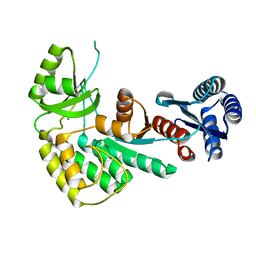 | |
2L4Z
 
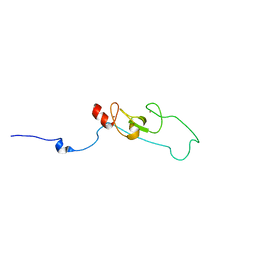 | | NMR structure of fusion of CtIP (641-685) to LMO4-LIM1 (18-82) | | Descriptor: | DNA endonuclease RBBP8,LIM domain transcription factor LMO4, ZINC ION | | Authors: | Liew, C, Stokes, P.H, Kwan, A.H, Matthews, J.M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction of the Breast Cancer Oncogene LMO4 with the Tumour Suppressor CtIP/RBBP8.
J.Mol.Biol., 425, 2013
|
|
1Y0J
 
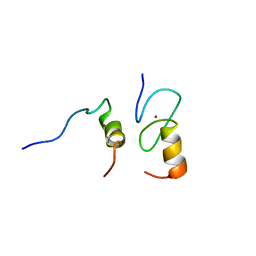 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4URS
 
 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
8IPP
 
 | | Crystal structure of the complex between an ankyrin and a parallel G-quadruplex | | Descriptor: | 93del-T12, DARPin protein 2E4, MAGNESIUM ION, ... | | Authors: | Ngo, K.H, Liew, C.W, Heddi, B, Phan, A.T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural Basis for Parallel G-Quadruplex Recognition by an Ankyrin Protein.
J.Am.Chem.Soc., 146, 2024
|
|
8IPF
 
 | |
5WYQ
 
 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
5WYR
 
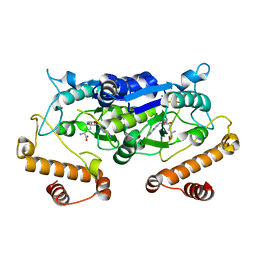 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
7XH9
 
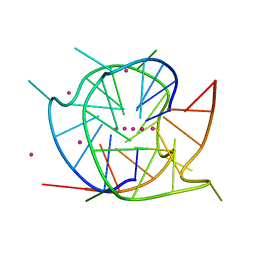 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*CP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION, STRONTIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7XHD
 
 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A•G•G•G•G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7XIE
 
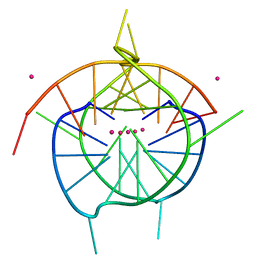 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*GP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A•G•G•G•G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
6LE9
 
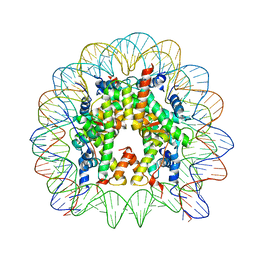 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-24 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6L9H
 
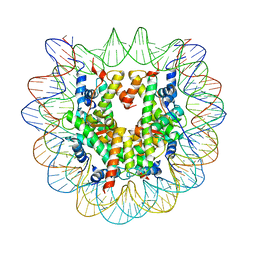 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
1M36
 
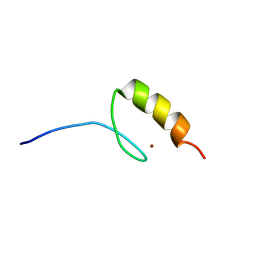 | |
7X7G
 
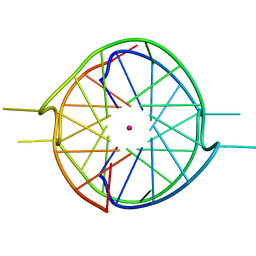 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
2HVA
 
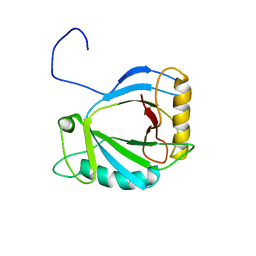 | | Solution Structure of the haem-binding protein p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Gell, D.A, Mackay, J.P, Westman, B.J, Liew, C.K, Gorman, D. | | Deposit date: | 2006-07-28 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Novel Haem-binding Interface in the 22 kDa Haem-binding Protein p22HBP.
J.Mol.Biol., 362, 2006
|
|
