5YAO
 
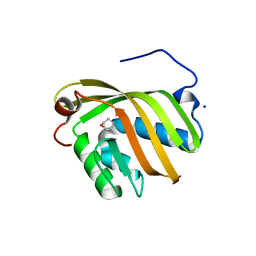 | | The complex structure of SZ529 and expoxid | | 分子名称: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, SODIUM ION | | 著者 | Lian, W, Sun, Z.T, Zhou, J.H, Reetz, M.T. | | 登録日 | 2017-09-01 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.611 Å) | | 主引用文献 | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations
J. Am. Chem. Soc., 140, 2018
|
|
5COR
 
 | |
5COY
 
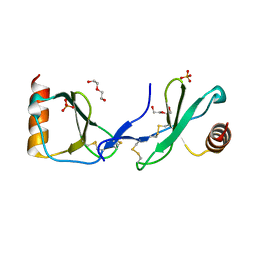 | | Crystal structure of CC chemokine 5 (CCL5) | | 分子名称: | C-C motif chemokine 5, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Liang, W.G, Tang, W. | | 登録日 | 2015-07-20 | | 公開日 | 2016-04-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.443 Å) | | 主引用文献 | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CMD
 
 | |
5CJO
 
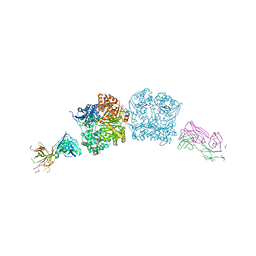 | |
5UOE
 
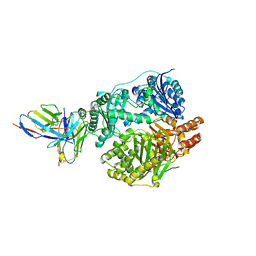 | |
6XOS
 
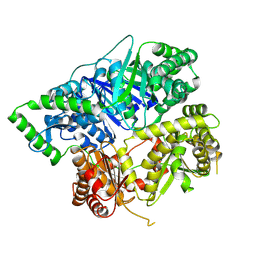 | |
6XOU
 
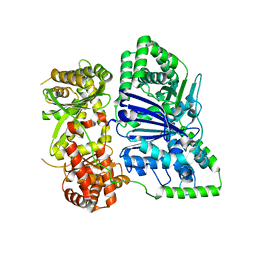 | |
6XOV
 
 | |
6XOW
 
 | |
6XOT
 
 | |
6XLY
 
 | |
6B70
 
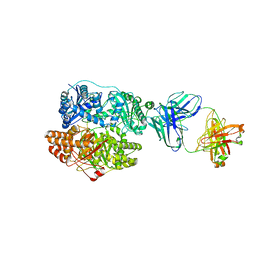 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | 分子名称: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-03 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF8
 
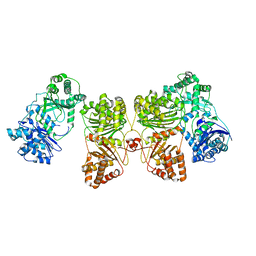 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Y
 
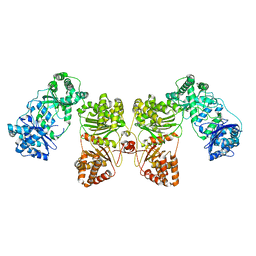 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BFC
 
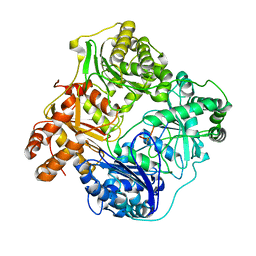 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF6
 
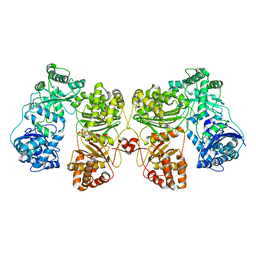 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B3Q
 
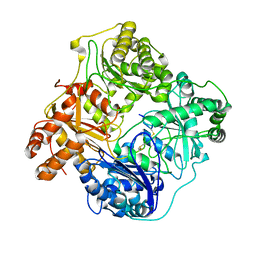 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-09-22 | | 公開日 | 2017-11-22 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Z
 
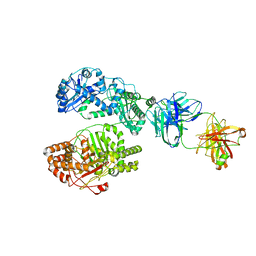 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | 分子名称: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2018-01-10 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
4RE9
 
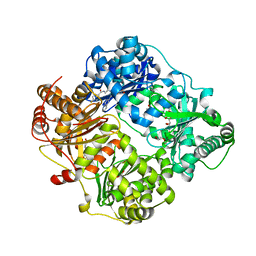 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | 著者 | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | 登録日 | 2014-09-22 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.908 Å) | | 主引用文献 | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4RA8
 
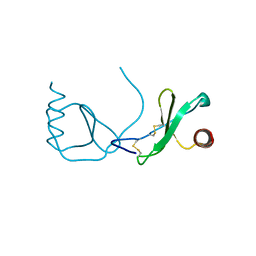 | | Structure analysis of the Mip1a P8A mutant | | 分子名称: | C-C motif chemokine 3 | | 著者 | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | 登録日 | 2014-09-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
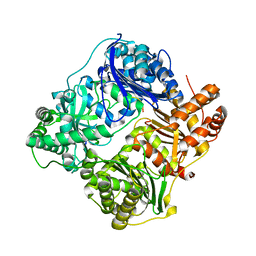 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | 分子名称: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | 著者 | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | 登録日 | 2014-09-10 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.148 Å) | | 主引用文献 | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
6C6D
 
 | |
