4RLT
 
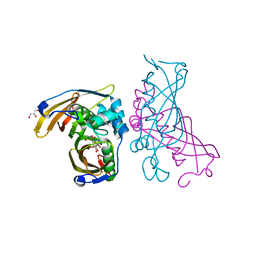 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Fisetin | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 3,7,3',4'-TETRAHYDROXYFLAVONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
5HQB
 
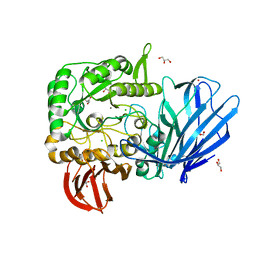 | | A Glycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
6MTG
 
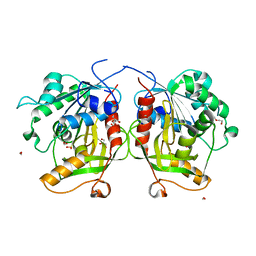 | | A Single Reactive Noncanonical Amino Acid is Able to Dramatically Stabilize Protein Structure | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, J.C, Nasertorabi, F, Xuan, W, Han, G.W, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2018-10-19 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Single Reactive Noncanonical Amino Acid Is Able to Dramatically Stabilize Protein Structure.
Acs Chem.Biol., 14, 2019
|
|
6WQX
 
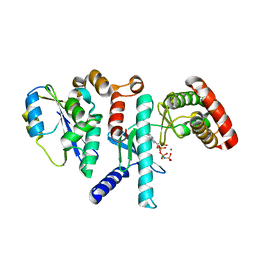 | | Human PRPK-TPRKB complex | | Descriptor: | EKC/KEOPS complex subunit TP53RK, EKC/KEOPS complex subunit TPRKB, MAGNESIUM ION, ... | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the human PRPK-TPRKB complex.
Commun Biol, 4, 2021
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
6DQ3
 
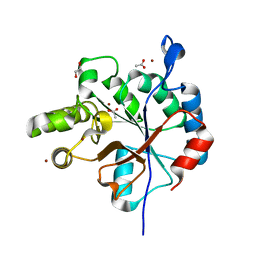 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
4BBE
 
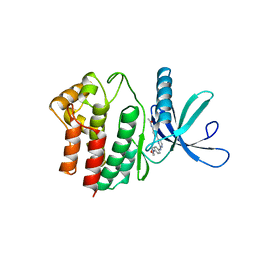 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]ethanamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4BBF
 
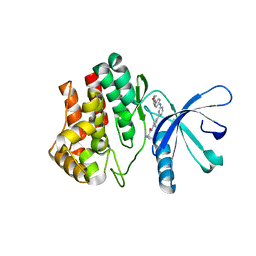 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | (2R)-N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]pyrrolidine-2-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4Y5U
 
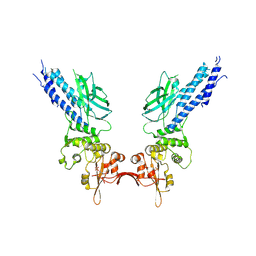 | | Transcription factor | | Descriptor: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4EFO
 
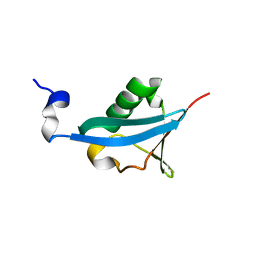 | | Crystal structure of the ubiquitin-like domain of human TBK1 | | Descriptor: | Serine/threonine-protein kinase TBK1 | | Authors: | Li, J, Li, J, Miyahira, A, Sun, J, Liu, Y, Cheng, G, Liang, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of the ubiquitin-like domain of human TBK1.
Protein Cell, 3, 2012
|
|
2ESY
 
 | | Structure and influence on stability and activity of the N-terminal propetide part of lung surfactant protein C | | Descriptor: | lung surfactant protein C | | Authors: | Li, J, Liepinsh, E, Almlen, A, Thyberg, J, Curstedt, T, Jornvall, H, Johansson, J. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure and influence on stability and activity of the N-terminal propeptide part of lung surfactant protein C
Febs J., 273, 2006
|
|
8TTF
 
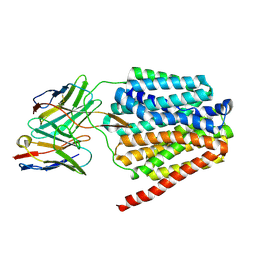 | | NorA double mutant - E222QD307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTH
 
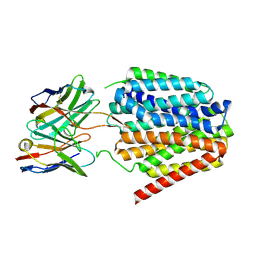 | | NorA single mutant - D307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTE
 
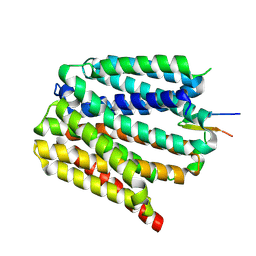 | | Protonated state of NorA at pH 5.0 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTG
 
 | | NorA single mutant - E222Q at pH 7.5 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
5D39
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
4GAX
 
 | | Crystal Structure of an alpha-Bisabolol synthase mutant | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Li, J, Peng, Z. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
4L8Z
 
 | | Crystal structure of Human Hsp90 with RL1 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](3,4-dihydroisoquinolin-2(1H)-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | Descriptor: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4L94
 
 | | Crystal structure of Human Hsp90 with S46 | | Descriptor: | (4-hydroxyphenyl)(4-methylpiperazin-1-yl)methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5CF0
 
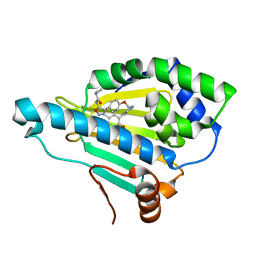 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(isoquinolin-4-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | FS23 binds to the N-terminal domain of human Hsp90: A novel small inhibitor for Hsp90
Nucl.Sci.Tech., 26, 2015
|
|
2AD6
 
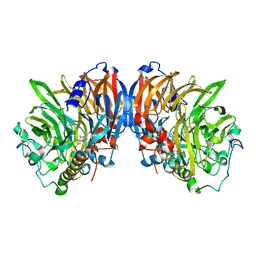 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
2AD8
 
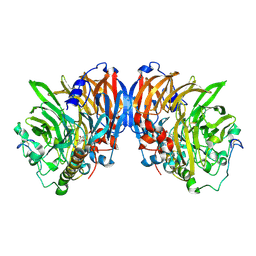 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) in the presence of ethanol | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
