3KAY
 
 | | Crystal structure of abscisic acid receptor PYL1 | | Descriptor: | Putative uncharacterized protein At5g46790 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB0
 
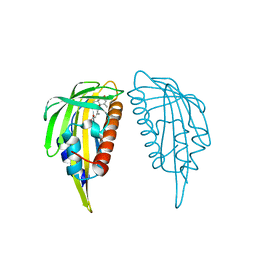 | | Crystal structure of abscisic acid-bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KAZ
 
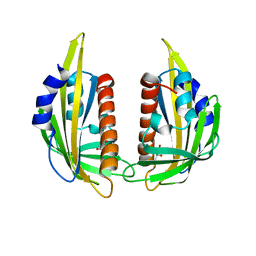 | | Crystal structure of abscisic acid receptor PYL2 | | Descriptor: | 1,3-BUTANEDIOL, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB3
 
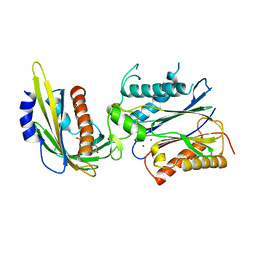 | | Crystal structure of abscisic acid-bound PYL2 in complex with HAB1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Melcher, K, Soon, F.-F, Ng, L.-M, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3COX
 
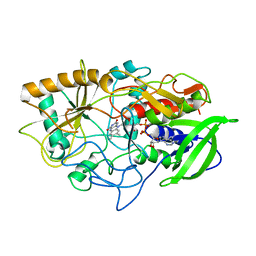 | | CRYSTAL STRUCTURE OF CHOLESTEROL OXIDASE COMPLEXED WITH A STEROID SUBSTRATE. IMPLICATIONS FOR FAD DEPENDENT ALCOHOL OXIDASES | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Vrielink, A, Li, J, Brick, P, Blow, D.M. | | Deposit date: | 1993-06-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cholesterol oxidase complexed with a steroid substrate: implications for flavin adenine dinucleotide dependent alcohol oxidases.
Biochemistry, 32, 1993
|
|
4S39
 
 | | IspG in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3C
 
 | | IspG in complex with Epoxide Intermediate | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3E
 
 | | IspG in complex with Inhibitor 7 (compound 1061) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
6XLT
 
 | |
4S3B
 
 | | IspG in complex with Intermediate II | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3D
 
 | | IspG in complex with PPi | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
6XLS
 
 | |
4S3F
 
 | | IspG in complex with Inhibitor 8 (compound 1077) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, FE3-S4 CLUSTER, but-3-yn-1-yl trihydrogen diphosphate | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3A
 
 | | IspG in complex with Intermediate I | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
7VQX
 
 | | Cryo-EM structure of human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-10-21 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
4HK4
 
 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
4HPW
 
 | | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine | | Descriptor: | 3-methoxy-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine
To be Published
|
|
4I42
 
 | | E.coli. 1,4-dihydroxy-2-naphthoyl coenzyme A synthase (ecMenB) in complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA, ... | | Authors: | Sun, Y, Song, H, Li, J, Li, Y, Jiang, M, Zhou, J, Guo, Z. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme A synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4ELW
 
 | | Structure of E. coli. 1,4-dihydroxy-2- naphthoyl coenzyme A synthases (MENB) in complex with nitrate | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sun, Y.R, Song, H.G, Li, J, Jiang, M, Li, Y, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-04-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways
Biochemistry, 51, 2012
|
|
4P5A
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
4P5B
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br dUMP | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB form Streptomyces cacaoi bound with 5-Br dUMP
To Be Published
|
|
3DC1
 
 | | Crystal structure of kynurenine aminotransferase II complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Kynurenine/alpha-aminoadipate aminotransferase mitochondrial | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
Biosci.Rep., 28, 2008
|
|
1DC2
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
4YHC
 
 | | Crystal structure of the WD40 domain of SCAP from fission yeast | | Descriptor: | CITRIC ACID, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Li, J.X, Wu, J.P, Yan, C.Y, Yan, N. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the WD40 domain of SCAP from fission yeast reveals the molecular basis for SREBP recognition.
Cell Res., 25, 2015
|
|
4ELS
 
 | | Structure of E. Coli. 1,4-dihydroxy-2- naphthoyl coenzyme A synthases (MENB) in complex with bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, BICARBONATE ION, ... | | Authors: | Sun, Y.R, Song, H.G, Li, J, Jiang, M, Li, Y, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-04-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways
Biochemistry, 51, 2012
|
|
