8CWS
 
 | |
8CUU
 
 | |
8CUV
 
 | |
8E2L
 
 | | Structure of Lates calcarifer Twinkle helicase with ATP and DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | 著者 | Gao, Y, Li, Z. | | 登録日 | 2022-08-15 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Structural and dynamic basis of DNA capture and translocation by mitochondrial Twinkle helicase.
Nucleic Acids Res., 50, 2022
|
|
4R05
 
 | | Crystal structure of the refolded DENV3 methyltransferase | | 分子名称: | Nonstructural protein NS5 | | 著者 | Brecher, M.B, Li, Z, Zhang, J, Chen, H, Lin, Q, Liu, B, Li, H.M. | | 登録日 | 2014-07-29 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase.
Protein Sci., 24, 2015
|
|
1M62
 
 | | Solution structure of the BAG domain from BAG4/SODD | | 分子名称: | BAG-family molecular chaperone regulator-4 | | 著者 | Briknarova, K, Takayama, S, Homma, S, Baker, K, Cabezas, E, Hoyt, D.W, Li, Z, Satterthwait, A.C, Ely, K.R. | | 登録日 | 2002-07-11 | | 公開日 | 2002-07-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | BAG4/SODD protein contains a short BAG domain.
J.Biol.Chem., 277, 2002
|
|
6M4P
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-bound structure | | 分子名称: | 6-methoxy-streptovaricin C, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | 登録日 | 2020-03-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
3KUP
 
 | | Crystal Structure of the CBX3 Chromo Shadow Domain | | 分子名称: | Chromobox protein homolog 3, UNKNOWN ATOM OR ION | | 著者 | Tempel, W, Li, Z, Li, Y, Kozieradzki, I, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-11-27 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal Structure of the CBX3 Chromo Shadow Domain
to be published
|
|
6AEI
 
 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | 著者 | Duan, J, Li, Z, Li, J, Zhang, J. | | 登録日 | 2018-08-05 | | 公開日 | 2019-08-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-04 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.75 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-04 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (6.75 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
8K98
 
 | |
8K9A
 
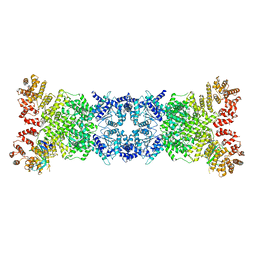 | | Cryo-EM structure of DSR2-DSAD1 state 2 | | 分子名称: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | 著者 | Zhang, H, Li, Z, Li, X.Z. | | 登録日 | 2023-07-31 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
6BWD
 
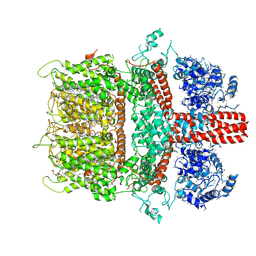 | | 3.7 angstrom cryoEM structure of truncated mouse TRPM7 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, MAGNESIUM ION, Transient receptor potential cation channel subfamily M member 7 | | 著者 | Zhang, J, Li, Z, Duan, J, Li, J, Hulse, R.E, Santa-Cruz, A, Abiria, S.A, Krapivinsky, G, Clapham, D.E. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BWI
 
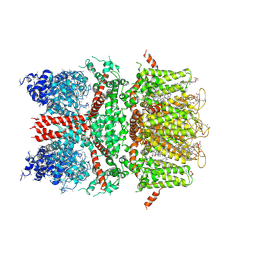 | | 3.7 angstrom cryoEM structure of full length human TRPM4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | 著者 | Zhang, J, Li, Z, Duan, J, Li, J, Clapham, D.E. | | 登録日 | 2017-12-15 | | 公開日 | 2018-12-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of full-length human TRPM4.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
5H4P
 
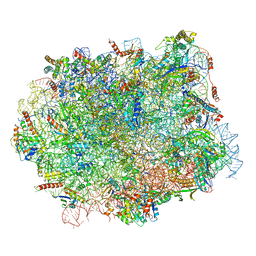 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | 登録日 | 2016-11-01 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
1DBX
 
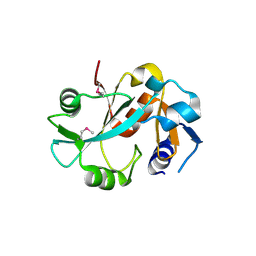 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase from H. influenzae (HI1434) | | 分子名称: | cysteinyl-tRNA(Pro) deacylase | | 著者 | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 1999-11-03 | | 公開日 | 2000-06-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | 著者 | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | 登録日 | 2015-03-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.916 Å) | | 主引用文献 | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
1FJ8
 
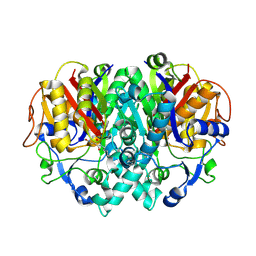 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH CERULENIN, IMPLICATIONS FOR DRUG DESIGN | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I | | 著者 | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | 登録日 | 2000-08-07 | | 公開日 | 2000-08-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
1DBU
 
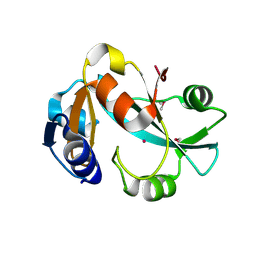 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | 分子名称: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | 著者 | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 1999-11-03 | | 公開日 | 2000-06-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
1FJ4
 
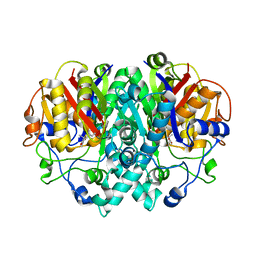 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH THIOLACTOMYCIN, IMPLICATIONS FOR DRUG DESIGN | | 分子名称: | BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I, THIOLACTOMYCIN | | 著者 | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | 登録日 | 2000-08-07 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
