3J6J
 
 | | 3.6 Angstrom resolution MAVS filament generated from helical reconstruction | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Wu, B, Peisley, A, Li, Z, Egelman, E, Walz, T, Penczek, P, Hur, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I.
Mol.Cell, 55, 2014
|
|
5A9V
 
 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A6F
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A9W
 
 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A22
 
 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | Descriptor: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | Authors: | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
5A6E
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, ... | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
6DU8
 
 | | Human Polycsytin 2-l1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystic kidney disease 2-like 1 protein | | Authors: | Hulse, R.E, Clapham, D.E, Li, Z, Huang, R.K, Zhang, J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of the polycystin 2-l1 ion channel.
Elife, 7, 2018
|
|
8J7Z
 
 | | Structure of FCP trimer in Cyclotella meneghiniana | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, CHLOROPHYLL A, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
5A6G
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, S1-S4 DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
6JBU
 
 | |
7KC2
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KCB
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NAD+ and Trifluoroethanol | | Descriptor: | ADH1 isoform 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KCQ
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Open Form of Apoenzyme | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7YNY
 
 | |
5F1M
 
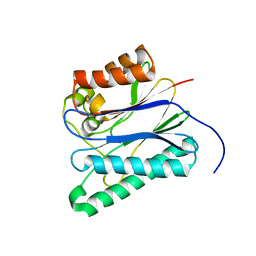 | |
5Y9F
 
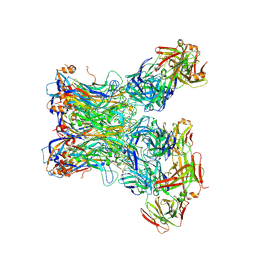 | | Crystal structure of HPV59 pentamer in complex with the Fab fragment of antibody 28F10 | | Descriptor: | Major capsid protein L1, heavy chain of Fab fragment of antibody 28F10, light chains of Fab fragment of antibody 28F10 | | Authors: | Li, S.W, Li, Z.H. | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.
MBio, 8, 2017
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
7CIH
 
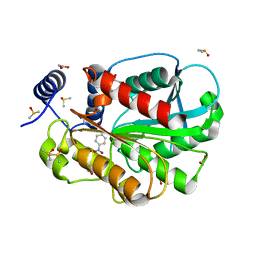 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
3MGM
 
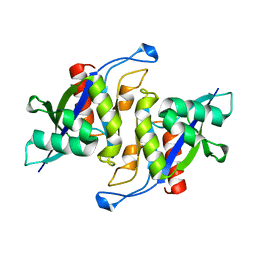 | | Crystal structure of human NUDT16 | | Descriptor: | U8 snoRNA-decapping enzyme | | Authors: | Yan, J, Lu, G, Zhang, J, Qi, J, Li, Z, Gao, F. | | Deposit date: | 2010-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure and the template mRNA decapping activity of human NUDT16
To be Published
|
|
8IL7
 
 | |
3OW8
 
 | | Crystal Structure of the WD repeat-containing protein 61 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 61 | | Authors: | Tempel, W, Li, Z, Chao, X, Lam, R, Wernimont, A.K, He, H, Seitova, A, Pan, P.W, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
3MHD
 
 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
1DBX
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase from H. influenzae (HI1434) | | Descriptor: | cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
