3B98
 
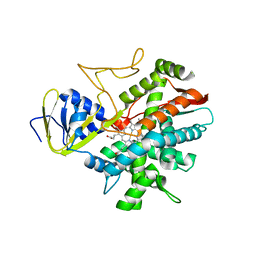 | | Crystal structure of zebrafish prostacyclin synthase (cytochrome P450 8A1) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostaglandin I2 synthase | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-11-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
3B6H
 
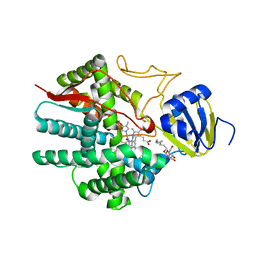 | | Crystal structure of human prostacyclin synthase in complex with inhibitor minoxidil | | Descriptor: | 6-PIPERIDIN-1-YLPYRIMIDINE-2,4-DIAMINE 3-OXIDE, PROTOPORPHYRIN IX CONTAINING FE, Prostacyclin synthase, ... | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
8JOL
 
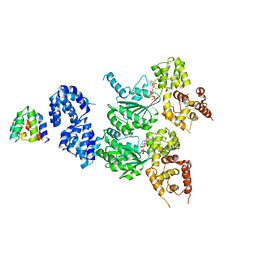 | | cryo-EM structure of the CED-4/CED-3 holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 3, Cell death protein 4, ... | | Authors: | Li, Y, Tian, L, Zhang, Y, Shi, Y. | | Deposit date: | 2023-06-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
5DO2
 
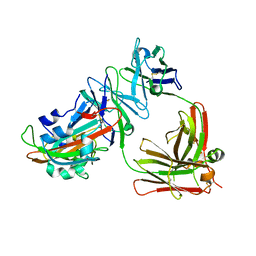 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
3FYH
 
 | | Recombinase in complex with ADP and metatungstate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2009-01-22 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal Rad51 homologue in complex with a metatungstate inhibitor.
Biochemistry, 48, 2009
|
|
7CYV
 
 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | Descriptor: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
3ETL
 
 | | RadA recombinase from Methanococcus maripaludis in complex with AMPPNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EWA
 
 | | RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and ammonium ions | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EW9
 
 | | RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and potassium ions | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4NHJ
 
 | | Crystal structure of Klebsiella pneumoniae RstA DNA-binding domain in complex with RstA box | | Descriptor: | 5'-D(*CP*AP*GP*GP*GP*AP*GP*TP*AP*AP*CP*GP*GP*AP*AP*TP*GP*TP*AP*CP*AP*AP*C)-3', 5'-D(*GP*GP*TP*TP*GP*TP*AP*CP*AP*TP*TP*CP*CP*GP*TP*TP*AP*CP*TP*CP*CP*CP*T)-3', DNA-binding transcriptional regulator RstA | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2013-11-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
3JBB
 
 | | Characterization of red-shifted phycobiliprotein complexes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris | | Descriptor: | PHYCOCYANOBILIN, SULFATE ION, allophycocyanin beta chain, ... | | Authors: | Li, Y, Lin, Y, Garvey, C, Birch, D, Corkery, R.W, Loughlin, P.C, Scheer, H, Willows, R.D, Chen, M. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-11 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Characterization of red-shifted phycobilisomes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris.
Biochim.Biophys.Acta, 1857, 2015
|
|
1KQL
 
 | | Crystal structure of the C-terminal region of striated muscle alpha-tropomyosin at 2.7 angstrom resolution | | Descriptor: | Fusion Protein of and striated muscle alpha-tropomyosin and the GCN4 leucine zipper | | Authors: | Li, Y, Mui, S, Brown, J.H, Strand, J, Reshetnikova, L, Tobacman, L.S, Cohen, C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4NIC
 
 | |
4YZE
 
 | |
8J4I
 
 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
4ZVV
 
 | | Lactate dehydrogenase A in complex with a trisubstituted piperidine-2,4-dione inhibitor GNE-140 | | Descriptor: | (2~{R})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Li, Y, Chen, Z, Eigenbrot, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition.
Nat.Chem.Biol., 12, 2016
|
|
3D34
 
 | | Structure of the F-spondin domain of mindin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Spondin-2 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-02-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule.
Embo J., 28, 2009
|
|
5ZN9
 
 | | Crystal structure of PX domain | | Descriptor: | SULFATE ION, Sorting nexin-27 | | Authors: | Li, Y, Zhu, Z, Li, F, Liao, S, Xu, C. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Crystal structure of PX domain
To Be Published
|
|
4P5B
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br dUMP | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB form Streptomyces cacaoi bound with 5-Br dUMP
To Be Published
|
|
3HHS
 
 | | Crystal Structure of Manduca sexta prophenoloxidase | | Descriptor: | COPPER (II) ION, Phenoloxidase subunit 1, Phenoloxidase subunit 2 | | Authors: | Li, Y, Wang, Y, Jiang, H, Deng, J. | | Deposit date: | 2009-05-17 | | Release date: | 2009-09-29 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8GJY
 
 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
4P5A
 
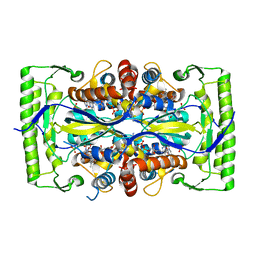 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
1KGD
 
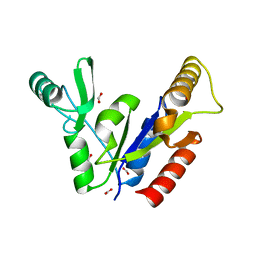 | | Crystal Structure of the Guanylate Kinase-like Domain of Human CASK | | Descriptor: | FORMIC ACID, PERIPHERAL PLASMA MEMBRANE CASK | | Authors: | Li, Y, Spangenberg, O, Paarmann, I, Konrad, M, Lavie, A. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Structural basis for nucleotide-dependent regulation of membrane-associated guanylate kinase-like domains.
J.Biol.Chem., 277, 2002
|
|
8K80
 
 | |
6M62
 
 | |
