1HQR
 
 | | CRYSTAL STRUCTURE OF A SUPERANTIGEN BOUND TO THE HIGH-AFFINITY, ZINC-DEPENDENT SITE ON MHC CLASS II | | Descriptor: | HLA-DR ALPHA CHAIN, HLA-DR BETA CHAIN, MYELIN BASIC PROTEIN, ... | | Authors: | Li, Y, Li, H, Dimasi, N, Schlievert, P, Mariuzza, R. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
Immunity, 14, 2001
|
|
4N4G
 
 | |
4N4I
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
1QSY
 
 | | DDATP-Trapped closed ternary complex of the large fragment of DNA Polymerase I from thermus aquaticus | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, 5'-D(*AP*TP*TP*GP*CP*GP*CP*CP*TP*P*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3', ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3FF8
 
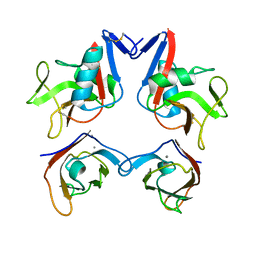 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | CALCIUM ION, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
1ZRN
 
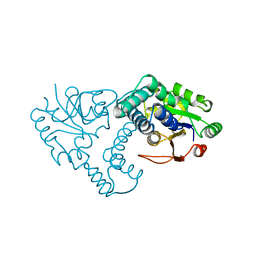 | | INTERMEDIATE STRUCTURE OF L-2-HALOACID DEHALOGENASE WITH MONOCHLOROACETATE | | Descriptor: | ACETIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
1ZRM
 
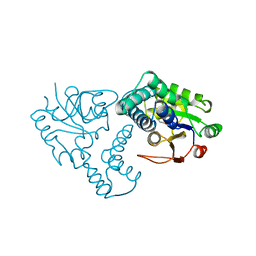 | | CRYSTAL STRUCTURE OF THE REACTION INTERMEDIATE OF L-2-HALOACID DEHALOGENASE WITH 2-CHLORO-N-BUTYRATE | | Descriptor: | L-2-HALOACID DEHALOGENASE, butanoic acid | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
1ZGL
 
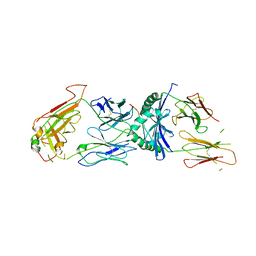 | | Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, Myelin basic protein, ... | | Authors: | Li, Y, Huang, Y, Lue, J, Quandt, J.A, Martin, R, Mariuzza, R.A. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
Embo J., 24, 2005
|
|
3FYH
 
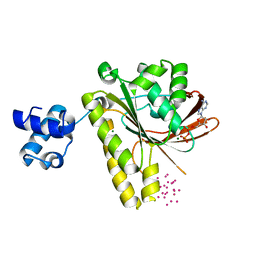 | | Recombinase in complex with ADP and metatungstate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2009-01-22 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal Rad51 homologue in complex with a metatungstate inhibitor.
Biochemistry, 48, 2009
|
|
8GJY
 
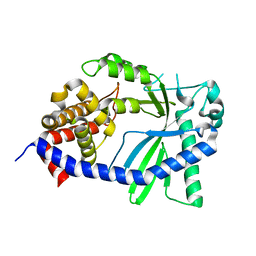 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
5Y27
 
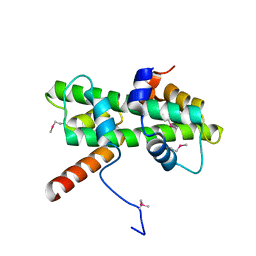 | | Crystal structure of Se-Met Dpb4-Dpb3 | | Descriptor: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | Authors: | Li, Y, Gao, F, Su, M, Zhang, F.B, Chen, Y.H. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8GJW
 
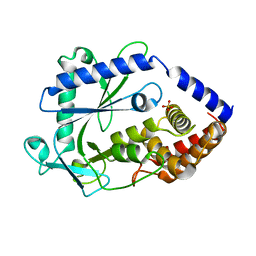 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJZ
 
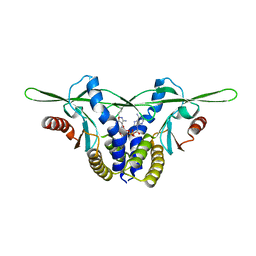 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
5YDM
 
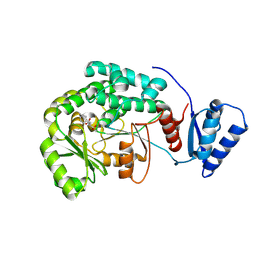 | |
6H8Q
 
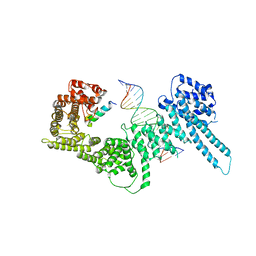 | | Structural basis for Scc3-dependent cohesin recruitment to chromatin | | Descriptor: | Cohesin subunit SCC3, DNA (5'-D(P*CP*TP*TP*TP*CP*GP*TP*TP*TP*CP*CP*TP*TP*GP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*CP*AP*AP*GP*GP*AP*AP*AP*CP*GP*AP*AP*AP*G)-3'), ... | | Authors: | Li, Y, Muir, K, Panne, D. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.631 Å) | | Cite: | Structural basis for Scc3-dependent cohesin recruitment to chromatin.
Elife, 7, 2018
|
|
9JN4
 
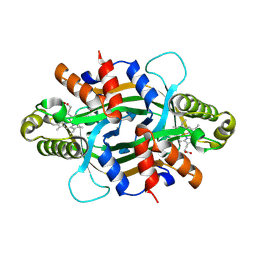 | | Crystal structure of KtzT-C197A in complex with HEME | | Descriptor: | FMN-binding protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
9JN6
 
 | | Crystal structure of KtzT-C197A in complex with HEME and product | | Descriptor: | (3~{S})-1,2-diazinane-3-carboxylic acid, FMN-binding protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
9JN5
 
 | | Crystal structure of KtzT-C197A in complex with HEME and substrate | | Descriptor: | FMN-binding protein, N~5~-hydroxy-L-ornithine, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
9KJR
 
 | |
9KJT
 
 | |
8T5K
 
 | | Crystal structure of STING CTD in complex with BDW-OH | | Descriptor: | Stimulator of interferon genes protein, {[(4S)-8,9-dimethylthieno[3,2-e][1,2,4]triazolo[4,3-c]pyrimidin-3-yl]sulfanyl}acetic acid | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
8T5L
 
 | | Crystal structure of STING CTD in complex with 2'3'-cGAMP | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
8ZXO
 
 | | Structure of the red fluorescent protein mScarlet-H at pH8.5 | | Descriptor: | red fluorescent protein mScarletH | | Authors: | Li, Y, Li, M, Fan, S, Zheng, Q, Li, S, Fu, Z. | | Deposit date: | 2024-06-14 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structure of the red fluorescent protein mScarlet-H at pH8.5
To Be Published
|
|
8S1R
 
 | |
8ZCJ
 
 | | Cryo-EM structure of the pasireotide-bound SSTR5-Gi complex | | Descriptor: | 004-DTR-LYS-TYR-PHA-HYP, Beta-2 adrenergic receptor,Somatostatin receptor type 5,lgbit (fusion protein), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y.G, Meng, X.Y, Yang, X.R, Ling, S.L, Shi, P, Tian, C.L, Yang, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into somatostatin receptor 5 bound with cyclic peptides.
Acta Pharmacol.Sin., 45, 2024
|
|
