7XST
 
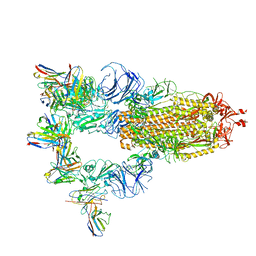 | |
7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7KHR
 
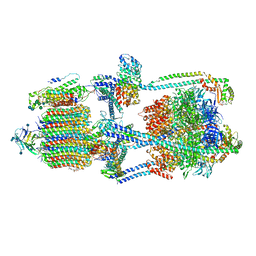 | | Cryo-EM structure of bafilomycin A1-bound intact V-ATPase from bovine brain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Molecular basis of V-ATPase inhibition by bafilomycin A1.
Nat Commun, 12, 2021
|
|
6GQM
 
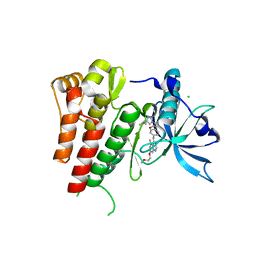 | | Crystal structure of human c-KIT kinase domain in complex with a small molecule inhibitor, AZD3229 | | Descriptor: | CHLORIDE ION, Mast/stem cell growth factor receptor Kit, ~{N}-[4-[[5-fluoranyl-7-(2-methoxyethoxy)quinazolin-4-yl]amino]phenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
5ZV7
 
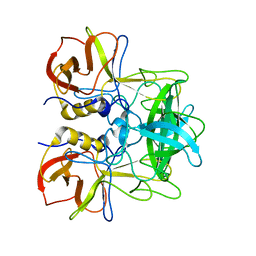 | | P domain of GII.17-2014/15 complexed with B-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZVC
 
 | | P domain of GII.13 norovirus capsid complexed with Lewis A trisaccharide | | Descriptor: | GLYCEROL, Major capsid protein VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZV9
 
 | | P domain of GII.13 norovirus capsid | | Descriptor: | GLYCEROL, Major capsid protein VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5HS5
 
 | |
5ZUQ
 
 | | P domain of GII.17-1978 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZUS
 
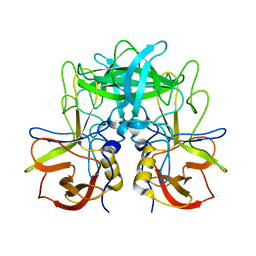 | | P domain of GII.17-2014/15 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZV5
 
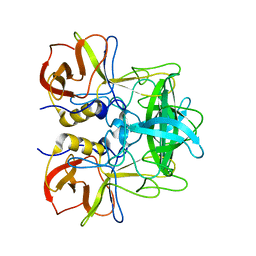 | | P domain of GII.17-2014/15 complexed with A-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5FO4
 
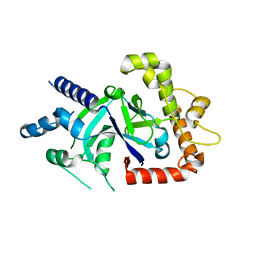 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P1) | | Descriptor: | LEUCYL TRNA SYNTHASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOC
 
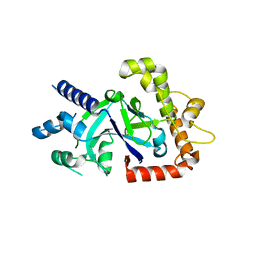 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P21) | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOD
 
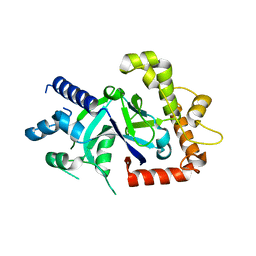 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P1) containing deletions of insertions 1 and 3 | | Descriptor: | 1,2-ETHANEDIOL, LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOF
 
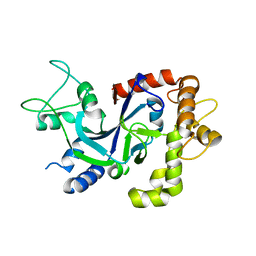 | | Crystal structure of the P.knowlesi cytosolic leucyl-tRNA synthetase editing domain | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5J08
 
 | |
6GQQ
 
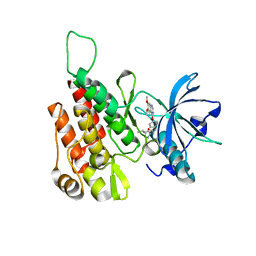 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 35) | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
3R27
 
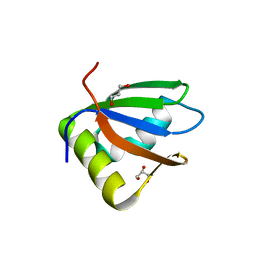 | | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L) | | Descriptor: | GLYCEROL, Heterogeneous nuclear ribonucleoprotein L | | Authors: | Zhang, W, Liu, Y, Zeng, F, Niu, L, Teng, M, Li, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L)
To be Published
|
|
5J8J
 
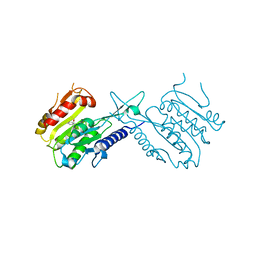 | | A histone deacetylase from Saccharomyces cerevisiae | | Descriptor: | Histone deacetylase HDA1 | | Authors: | Zhu, Y, Shen, H, Li, X, Teng, M. | | Deposit date: | 2016-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Structural and histone binding ability characterization of the ARB2 domain of a histone deacetylase Hda1 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
6GQL
 
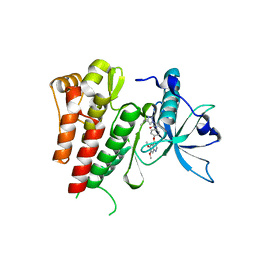 | | Crystal structure of human c-KIT kinase domain in complex with AZD3229-analogue (compound 35) | | Descriptor: | Mast/stem cell growth factor receptor Kit, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
6GQP
 
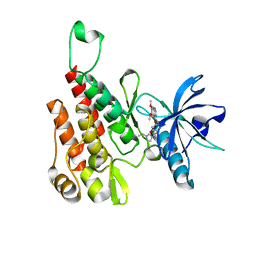 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 23) | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(1-ethylpyrazol-4-yl)ethanamide | | Authors: | Hardy, C.J, Schimpl, M, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
