2G5P
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 21ac | | Descriptor: | 4-{[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHOXY}-3-TERT-BUTYLBENZOIC ACID, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
2G63
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 24b | | Descriptor: | Dipeptidyl peptidase 4, METHYL 4-{[({[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHYL}AMINO)CARBONYL]AMINO}BENZOATE | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-24 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
4LZO
 
 | | Crystal structure of human PRS1 A87T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
8J85
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
4LYG
 
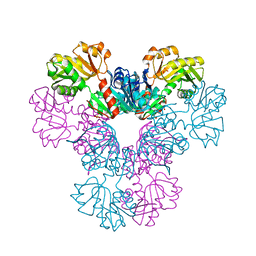 | | Crystal structure of human PRS1 E43T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4LZN
 
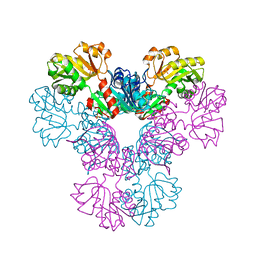 | | Crystal structure of human PRS1 D65N mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4M0P
 
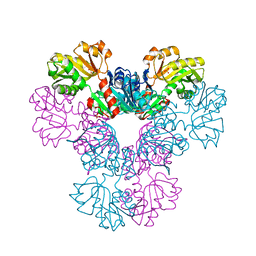 | | Crystal structure of human PRS1 M115T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4M0U
 
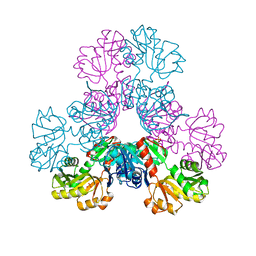 | | crystal structure of human PRS1 Q133P mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
8JCB
 
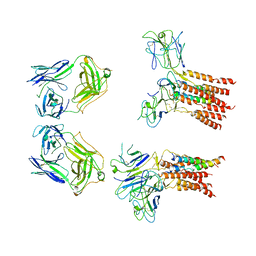 | | Vgamma5 Vdelta1 T cell receptor complex | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 2024
|
|
8JC0
 
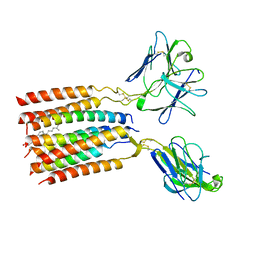 | | V gamma9 V delta2 TCR and CD3 complex in LMNG | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 2024
|
|
5X0X
 
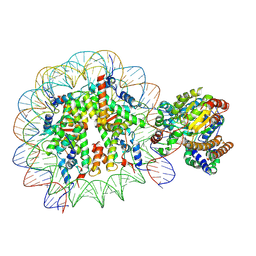 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
5X0Y
 
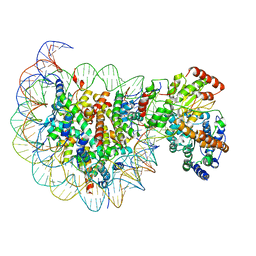 | | Complex of Snf2-Nucleosome complex with Snf2 bound to SHL2 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
4LA1
 
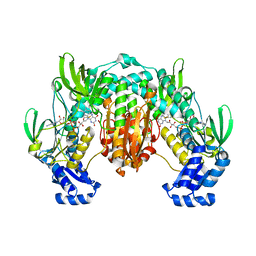 | | Crystal structure of SjTGR (thioredoxin glutathione reductase from Schistosoma japonicumi)complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Peng, Y, Wu, Q, Huang, F, Chen, J, Li, X, Zhou, X, Fan, X. | | Deposit date: | 2013-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of SjTGR complex with FAD
To be Published
|
|
3TO8
 
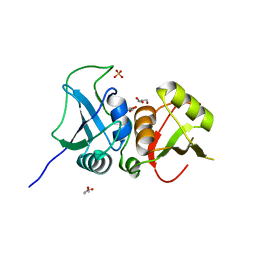 | | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Heterogeneous nuclear ribonucleoprotein L, ... | | Authors: | Zhang, W.J, Zeng, F.X, Liu, Y.W, Zhao, Y, Niu, L.W, Teng, M.K, Li, X. | | Deposit date: | 2011-09-04 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L)
To be Published
|
|
3VBC
 
 | |
6E1H
 
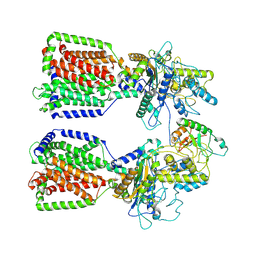 | | Structure of 2:1 human Ptch1-Shh-N complex | | Descriptor: | CALCIUM ION, PALMITIC ACID, Protein patched homolog 1, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex.
Science, 362, 2018
|
|
4KDC
 
 | | Crystal Structure of UBIG | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins.
Biochem. J., 470, 2015
|
|
4Q77
 
 | | Crystal structure of Rot, a global regulator of virulence genes in Staphylococcus aureus | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator rot | | Authors: | Zhu, Y, Fan, X, Li, X, Teng, M. | | Deposit date: | 2014-04-24 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of Rot, a global regulator of virulence genes in Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4A1G
 
 | | The crystal structure of the human Bub1 TPR domain in complex with the KI motif of Knl1 | | Descriptor: | MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, PROTEIN CASC5 | | Authors: | Krenn, V, Wehenkel, A, Li, X, Santaguida, S, Musacchio, A. | | Deposit date: | 2011-09-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis Reveals Features of the Spindle Checkpoint Kinase Bub1-Kinetochore Subunit Knl1 Interaction.
J.Cell Biol., 196, 2012
|
|
8CUF
 
 | | Synthetic epi-Novo29 (2R,3S), X-ray diffractometer structure | | Descriptor: | ACETATE ION, IODIDE ION, Synthetic epi-Novo29 (2R,3S) | | Authors: | Kreutzer, A.G, Li, X, Krumberger, M, Nowick, J.S. | | Deposit date: | 2022-05-17 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Synthesis and Stereochemical Determination of the Peptide Antibiotic Novo29.
J.Org.Chem., 88, 2023
|
|
8CUG
 
 | | Synthetic epi-Novo29 (2R,3S), synchrotron structure | | Descriptor: | ACETATE ION, Synthetic epi-Novo29 (2R,3S) | | Authors: | Kreutzer, A.G, Li, X, Krumberger, M, Nowick, J.S. | | Deposit date: | 2022-05-17 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Synthesis and Stereochemical Determination of the Peptide Antibiotic Novo29.
J.Org.Chem., 88, 2023
|
|
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
4D1Q
 
 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
8IGT
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A1 | | Descriptor: | Butyrophilin subfamily 2 member A1, ZINC ION | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2023-02-21 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
4DM4
 
 | | The conserved domain of yeast Cdc73 | | Descriptor: | Cell division control protein 73 | | Authors: | Chen, H, Shi, N, Gao, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
