7DBI
 
 | |
7DBL
 
 | |
7CAL
 
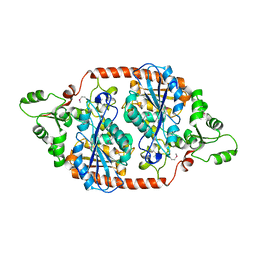
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | 著者 | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | 登録日 | 2020-06-09 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
8GUE
 
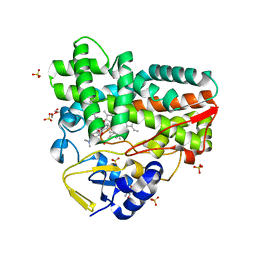 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | 著者 | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | 登録日 | 2022-09-11 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
6M4S
 
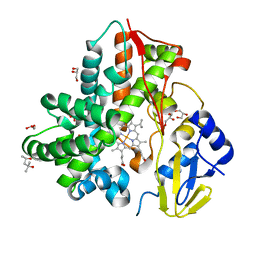 | | Crystal Structure Analysis of the cytochrome P450 CYP-Sb21 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cytochrome P450 hydroxylase sb21, ... | | 著者 | Li, F.W, Li, S.Y. | | 登録日 | 2020-03-09 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-guided manipulation of the regioselectivity of the cyclosporine A hydroxylase CYP-sb21 from Sebekia benihana .
Synth Syst Biotechnol, 5, 2020
|
|
7XBO
 
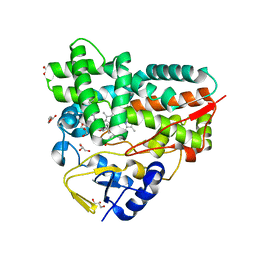 | | Crystal Structure of 10-dml-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | 分子名称: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | 登録日 | 2022-03-21 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBN
 
 | | Crystal Structure of YC-17-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | 分子名称: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | 登録日 | 2022-03-21 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBM
 
 | | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | 分子名称: | CACODYLATE ION, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | 登録日 | 2022-03-21 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238
Nat Commun, 2023
|
|
8XTF
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP-3C | | 分子名称: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide-3C, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | 登録日 | 2024-01-10 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTG
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and DMMPA | | 分子名称: | O-desmethyl mycophenolic acid, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | 登録日 | 2024-01-10 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTE
 
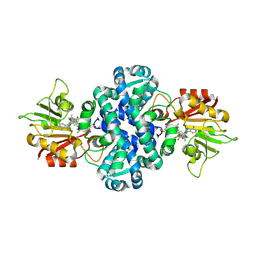 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP | | 分子名称: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | 登録日 | 2024-01-10 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
5YSW
 
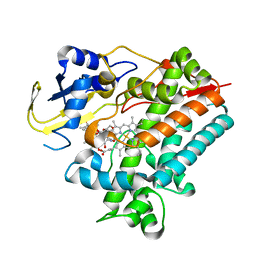 | | Crystal Structure Analysis of Rif16 in complex with R-L | | 分子名称: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | 登録日 | 2017-11-15 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5YSM
 
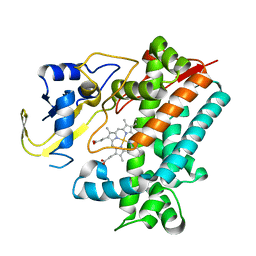 | | Crystal Structure Analysis of Rif16 | | 分子名称: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | 登録日 | 2017-11-14 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
7CCF
 
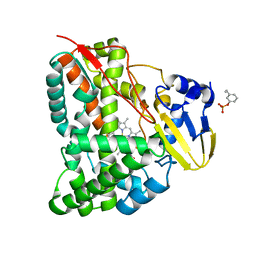 | | Mechanism insights on steroselective oxidation of phosphorylated ethylphenols with cytochrome P450 CreJ | | 分子名称: | (3-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Dong, S, Du, L, Li, S.Y, Feng, Y.G. | | 登録日 | 2020-06-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis for Selective Oxidation of Phosphorylated Ethylphenols by Cytochrome P450 Monooxygenase CreJ.
Appl.Environ.Microbiol., 87, 2021
|
|
7CFS
 
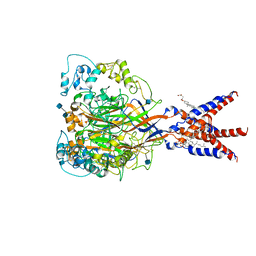 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | 登録日 | 2020-06-28 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7CFT
 
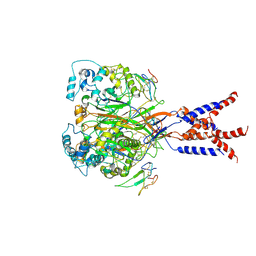 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | 著者 | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | 登録日 | 2020-06-28 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
