7L5X
 
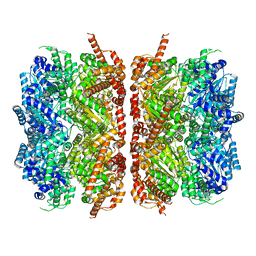 | | p97-R155H mutant dodecamer II | | 分子名称: | Transitional endoplasmic reticulum ATPase | | 著者 | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | 登録日 | 2020-12-23 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
3KS4
 
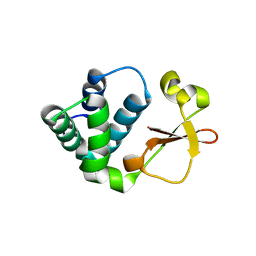 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain | | 分子名称: | Polymerase cofactor VP35 | | 著者 | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | 登録日 | 2009-11-20 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
5WPR
 
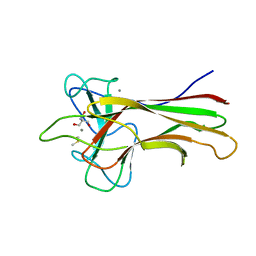 | | Crystal structure HpiC1 in C2 space group | | 分子名称: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5I72
 
 | | Crystal structure of the oligomeric form of the Lassa virus matrix protein Z | | 分子名称: | RING finger protein Z, ZINC ION | | 著者 | Hastie, K, Zandonatti, M, Liu, T, Li, S, Woods Jr, V, Saphire, E.O. | | 登録日 | 2016-02-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z.
J.Virol., 90, 2016
|
|
5X6U
 
 | | Crystal structure of human heteropentameric complex | | 分子名称: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | 著者 | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | 登録日 | 2017-02-23 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
5X6V
 
 | | Crystal structure of human heteroheptameric complex | | 分子名称: | ACETATE ION, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | 著者 | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | 登録日 | 2017-02-23 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
4GXB
 
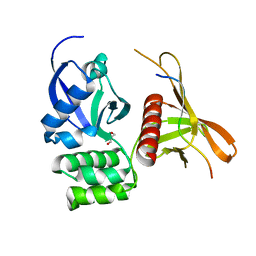 | | Structure of the SNX17 atypical FERM domain bound to the NPxY motif of P-selectin | | 分子名称: | GLYCEROL, P-selectin, Sorting nexin-17 | | 著者 | Ghai, R, Bugarcic, A, Liu, H, Norwood, S.J, Li, S.S, Teasdale, R.D, Collins, B.M. | | 登録日 | 2012-09-04 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5HYZ
 
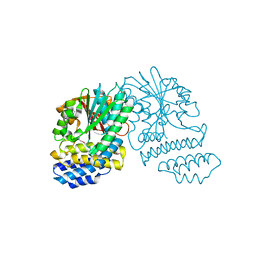 | | Crystal Structure of SCL7 in Oryza sativa | | 分子名称: | GRAS family transcription factor containing protein, expressed | | 著者 | Wu, Y, Li, S, Zhao, Y, Sun, L. | | 登録日 | 2016-02-02 | | 公開日 | 2016-04-27 | | 最終更新日 | 2016-06-29 | | 実験手法 | X-RAY DIFFRACTION (1.822 Å) | | 主引用文献 | Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa.
Plant Cell, 28, 2016
|
|
5WPS
 
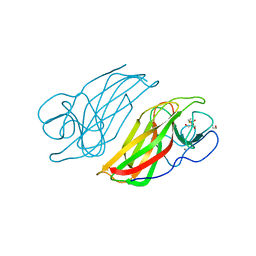 | | Crystal structure HpiC1 Y101F | | 分子名称: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.389 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
3KS8
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | 分子名称: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | 著者 | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | 登録日 | 2009-11-20 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
5WPU
 
 | | Crystal structure HpiC1 Y101S | | 分子名称: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
2AWT
 
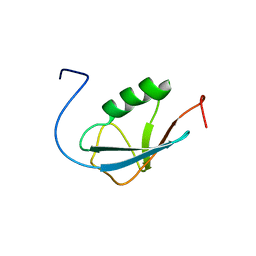 | | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2) | | 分子名称: | Small ubiquitin-related modifier 2 | | 著者 | Chang, C.K, Wang, Y.H, Chung, T.L, Chang, C.F, Li, S.S.L, Huang, T.H. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2)
To be Published
|
|
8JEU
 
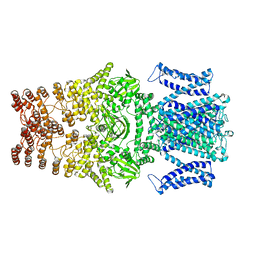 | |
8JEC
 
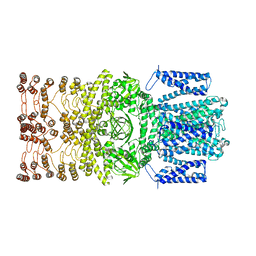 | |
8JET
 
 | |
6KTC
 
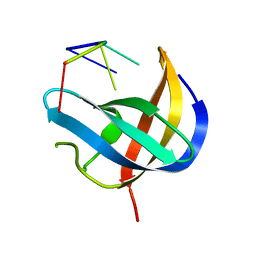 | | Crystal structure of YBX1 CSD with m5C RNA | | 分子名称: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*GP*(5MC)P*CP*U)-3') | | 著者 | Zou, F, Li, S. | | 登録日 | 2019-08-27 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KUG
 
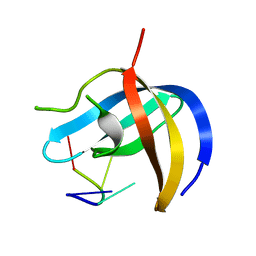 | | Crystal structure of YBX1 CSD with RNA | | 分子名称: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*GP*CP*CP*U)-3') | | 著者 | Zou, F, Li, S. | | 登録日 | 2019-09-02 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7VF5
 
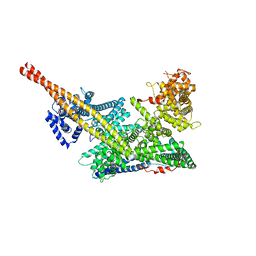 | | Human m6A-METTL associated complex (WTAP, VIRMA, and HAKAI) | | 分子名称: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog | | 著者 | Su, S, Li, S, Deng, T, Gao, M, Yin, Y, Wu, B, Peng, C, Liu, J, Ma, J, Zhang, K. | | 登録日 | 2021-09-10 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures of human m6A writer complexes.
Cell Res., 32, 2022
|
|
7VF2
 
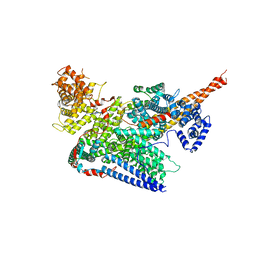 | | Human m6A-METTL associated complex (WTAP, VIRMA, ZC3H13, and HAKAI) | | 分子名称: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog, Zinc finger CCCH domain-containing protein 13 | | 著者 | Su, S, Li, S, Deng, T, Gao, M, Yin, Y, Wu, B, Peng, C, Liu, J, Ma, J, Zhang, K. | | 登録日 | 2021-09-10 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures of human m6A writer complexes.
Cell Res., 32, 2022
|
|
7WP1
 
 | | Cryo-EM structure of SARS-CoV-2 Mu S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WON
 
 | | Cryo-EM structure of SARS-CoV-2 S2P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP2
 
 | | Cryo-EM structure of SARS-CoV-2 C.1.2 S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP0
 
 | | Cryo-EM structure of SARS-CoV-2 Delta S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c335_light_IGLV1-40_IGLJ3, Spike protein S1, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
