8XFF
 
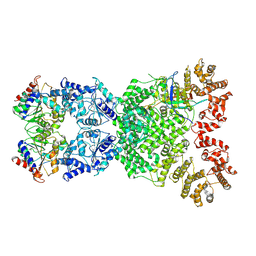 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein in complex with SPR phage tail tube protein | | Descriptor: | DSR2(H171A), SPR tail tube protein | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
5D1K
 
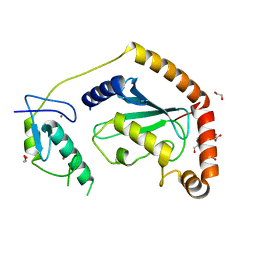 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
4HPH
 
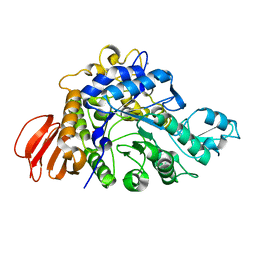 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOW
 
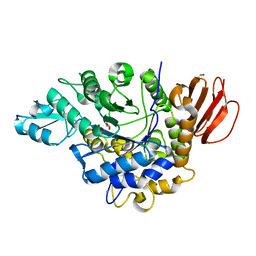 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOZ
 
 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
3QYC
 
 | | Structure of a dimeric anti-HER2 single domain antibody | | Descriptor: | VH domain of IgG molecule | | Authors: | Baral, T.N, Chao, S, Li, S, Tanha, J, Arbabai, M, Wang, S, Zhang, J. | | Deposit date: | 2011-03-03 | | Release date: | 2012-02-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Human Single Domain Antibody Dimer Formed through V(H)-V(H) Non-Covalent Interactions.
Plos One, 7, 2012
|
|
5TGZ
 
 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
4IH1
 
 | | Crystal structure of Karrikin Insensitive 2 (KAI2) from Arabidopsis thaliana | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH9
 
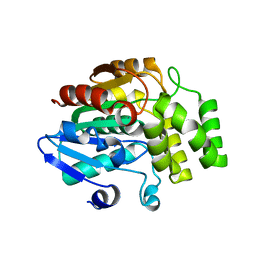 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
7MDO
 
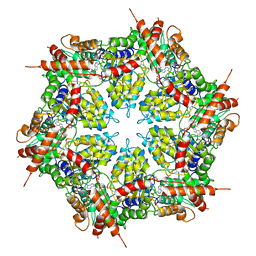 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
7MDM
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
6UET
 
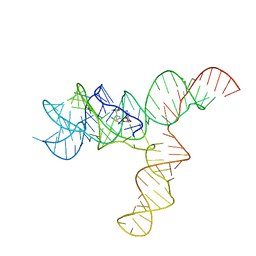 | | SAM-bound SAM-IV riboswitch | | Descriptor: | RNA (119-MER), S-ADENOSYLMETHIONINE | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6UES
 
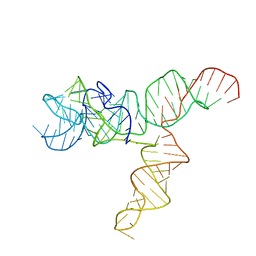 | | Apo SAM-IV Riboswitch | | Descriptor: | RNA (119-MER) | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
8GTD
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C portal-adaptor complex | | Descriptor: | Head-to-tail joining protein, Portal protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTF
 
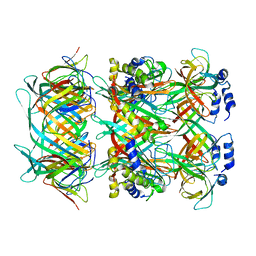 | | Cryo-EM model of the marine siphophage vB_DshS-R4C stopper-terminator complex | | Descriptor: | Head-to-tail joining protein, Major tail protein, Terminator protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTB
 
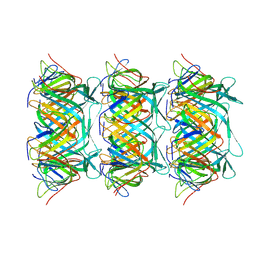 | | Cryo-EM structure of the marine siphophage vB_DshS-R4C tail tube protein | | Descriptor: | Major tail protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTC
 
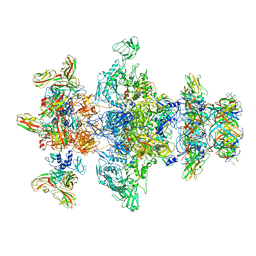 | | Cryo-EM model of the marine siphophage vB_DshS-R4C baseplate-tail complex | | Descriptor: | Distal tail protein, Hub protein, Major tail protein, ... | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
4IHA
 
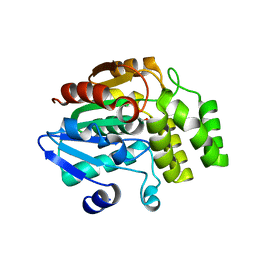 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
6V5C
 
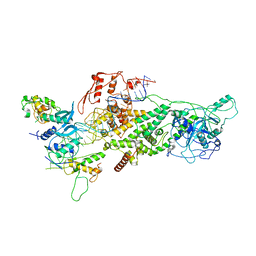 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
4IH4
 
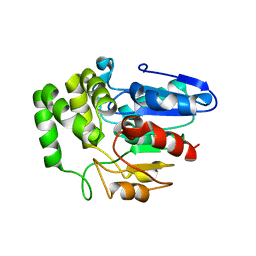 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | Descriptor: | AT3g03990/T11I18_10 | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
7Y53
 
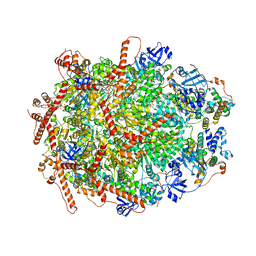 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
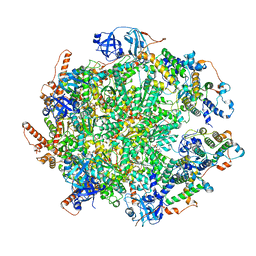 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
6V5B
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | Descriptor: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
