1HBY
 
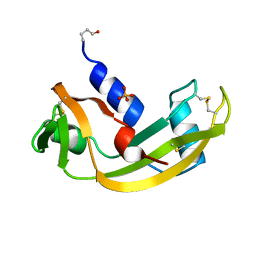 | | Binding of Phosphate and Pyrophosphate ions at the active site of human angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.S, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-04-21 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
6LK6
 
 | | MLKL mutant - T357AS358A | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
6M52
 
 | | Human apo ferritin frozen on TEM grid with amorphous carbon supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
1T7S
 
 | | Structural Genomics of Caenorhabditis elegans: Structure of BAG-1 protein | | Descriptor: | BAG-1 cochaperone | | Authors: | Symersky, J, Zhang, Y, Schormann, N, Li, S, Bunzel, R, Pruett, P, Luan, C.-H, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of the BAG domain.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6NUD
 
 | | Small conformation of ssRNA-bound CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
6NUE
 
 | | Small conformation of apo CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
6K7P
 
 | | Crystal structure of human AFF4-THD domain | | Descriptor: | AF4/FMR2 family member 4 | | Authors: | Tang, D, Xue, Y, Li, S, Cheng, W, Duan, J, Wang, J, Qi, S. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into the effect of AFF4 dimerization on activation of HIV-1 proviral transcription.
Cell Discov, 6, 2020
|
|
1YOP
 
 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
1M9C
 
 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-28 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
6KTC
 
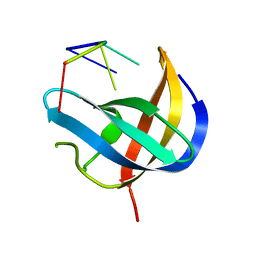 | | Crystal structure of YBX1 CSD with m5C RNA | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*GP*(5MC)P*CP*U)-3') | | Authors: | Zou, F, Li, S. | | Deposit date: | 2019-08-27 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1M9E
 
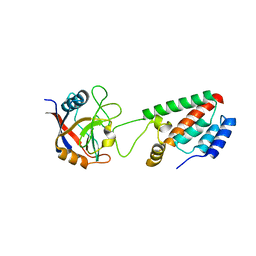 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-28 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
6KUG
 
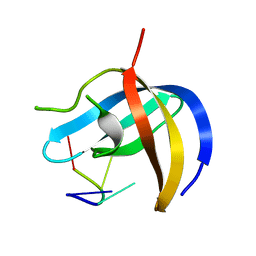 | | Crystal structure of YBX1 CSD with RNA | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*GP*CP*CP*U)-3') | | Authors: | Zou, F, Li, S. | | Deposit date: | 2019-09-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1M9X
 
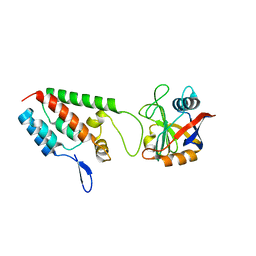 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,A88M,G89A Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-30 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
1M9Y
 
 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,G89A Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-30 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
4QEP
 
 | | crystal structure of KRYPTONITE in complex with mCHG DNA and SAH | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
4QEN
 
 | | crystal structure of KRYPTONITE in complex with mCHH DNA and SAH | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*TP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
4BLG
 
 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
4QEO
 
 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | Descriptor: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
3KS8
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
7YC8
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (388-MER) | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCI
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (389-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-2 RNA SL5 domain. | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7YCG
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
