5WIK
 
 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIJ
 
 | | JAK2 Pseudokinase in complex with NU6140 | | Descriptor: | 4-{[6-(cyclohexylmethoxy)-7H-purin-2-yl]amino}-N,N-diethylbenzamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIL
 
 | | JAK2 Pseudokinase in complex with AZD7762 | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
8GO3
 
 | |
8H8F
 
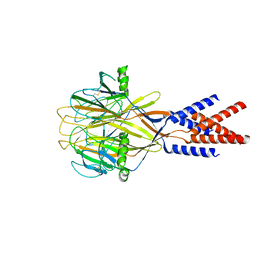 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state)
To Be Published
|
|
8H8E
 
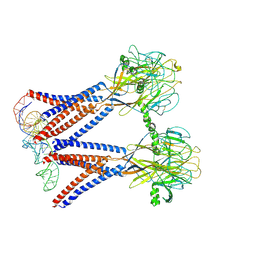 | | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state) | | Descriptor: | Proton-activated chloride channel, tRNA (75-MER)of Spodoptera frugiperda | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state)
To Be Published
|
|
8H8D
 
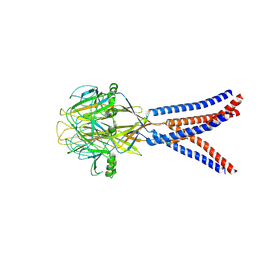 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state)
To Be Published
|
|
3HAG
 
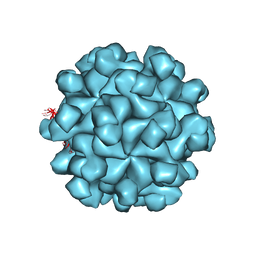 | | Crystal structure of the Hepatitis E Virus-like Particle | | Descriptor: | Capsid protein | | Authors: | Guu, T.S.Y, Liu, Z, Ye, Q, Mata, D.A, Li, K, Yin, C, Zhang, J, Tao, Y.J. | | Deposit date: | 2009-05-01 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5ISX
 
 | |
3JBM
 
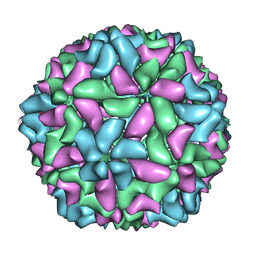 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
5ISW
 
 | |
3UTC
 
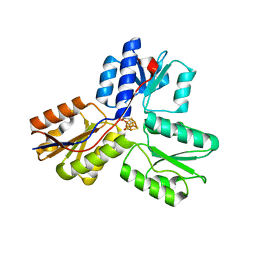 | | Ec_IspH in complex with (E)-4-hydroxybut-3-enyl diphosphate | | Descriptor: | (3E)-4-hydroxybut-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Li, K, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3MIL
 
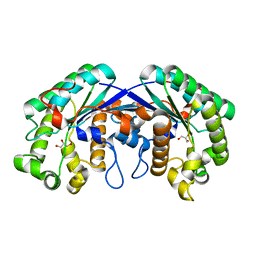 | | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Isoamyl acetate-hydrolyzing esterase | | Authors: | Ma, J, Lu, Q, Yuan, Y, Li, K, Ge, H, Go, Y, Niu, L, Teng, M. | | Deposit date: | 2010-04-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity
Proteins, 79, 2011
|
|
5K3H
 
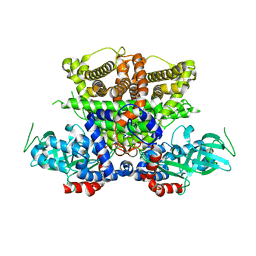 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-II | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZYO
 
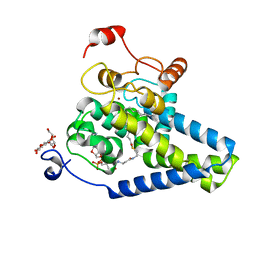 | | Crystal Structure of Human Integral Membrane Stearoyl-CoA Desaturase with Substrate | | Descriptor: | Acyl-CoA desaturase, DODECYL-BETA-D-MALTOSIDE, STEAROYL-COENZYME A, ... | | Authors: | Wang, H, Klein, M.G, Lane, W, Snell, G, Levin, I, Li, K, Zou, H, Sang, B.-C. | | Deposit date: | 2015-05-21 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5K3I
 
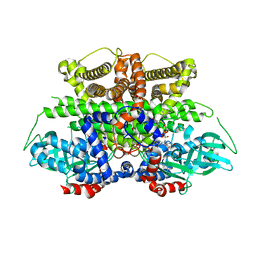 | | Crystal structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans complexed with FAD and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3G
 
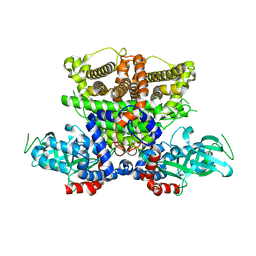 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-I | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8IVZ
 
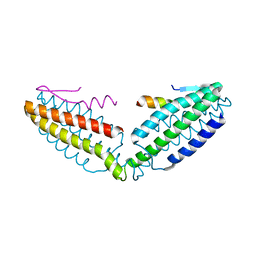 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | Descriptor: | KN motif and ankyrin repeat domains 1, Talin-1 | | Authors: | Xu, Y, Li, K, Wei, Z, Cong, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
5EG8
 
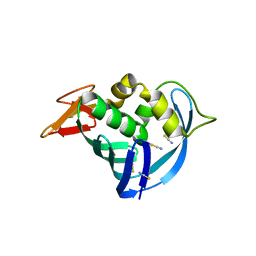 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2, THIOCYANATE ION | | Authors: | Liu, Y, Zheng, X, Severin, C, Rocha de Moura, T, Li, K, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EG7
 
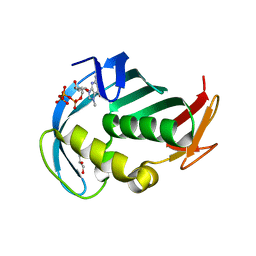 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EG9
 
 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4EA1
 
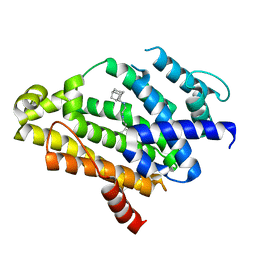 | | Co-crystal structure of dehydrosqualene synthase (Crtm) from S. aureus with SQ-109 | | Descriptor: | Dehydrosqualene synthase, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-N'-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]ethane-1,2-diamine | | Authors: | Lin, F.-Y, Li, K, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-03-21 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Head-to-Head Prenyl Tranferases: Anti-Infective Drug Targets.
J.Med.Chem., 55, 2012
|
|
8CXF
 
 | |
8DHC
 
 | |
