4R7A
 
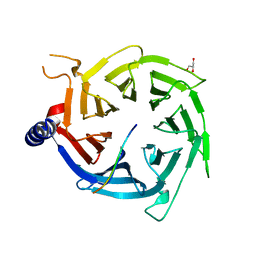 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
6B7N
 
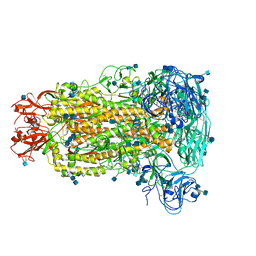 | | Cryo-electron microscopy structure of porcine delta coronavirus spike protein in the pre-fusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Tai, W, Du, L, Zhou, Y, Zhang, W, Li, F. | | Deposit date: | 2017-10-04 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy Structure of Porcine Deltacoronavirus Spike Protein in the Prefusion State
J. Virol., 92, 2018
|
|
2LS4
 
 | |
2LS2
 
 | |
4YND
 
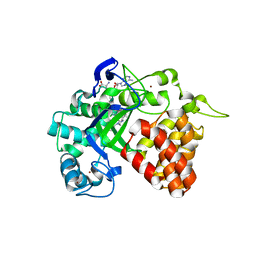 | | The Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2 | | Descriptor: | N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[(2R)-2-hydroxy-2-(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Sweis, R.F, Wang, Z, Algire, M, Arrowsmith, C.H, Brown, P.J, Chiang, G.C, Guo, J, Jakob, C.G, Kennedy, S, Li, F, Soni, N.B, Vedadi, M, Pappano, W.N. | | Deposit date: | 2015-03-09 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2.
Acs Med.Chem.Lett., 6, 2015
|
|
3CZ3
 
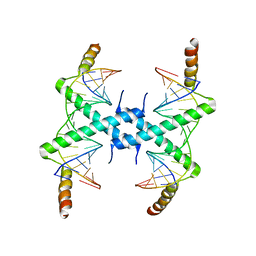 | | Crystal structure of Tomato Aspermy Virus 2b in complex with siRNA | | Descriptor: | Protein 2b, RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*A)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*G)-3') | | Authors: | Ma, J.B, Li, F, Ding, S.W, Patel, D.J. | | Deposit date: | 2008-04-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural Basis for siRNA Recognition by 2b, a Viral Suppressor of Non-Cell Autonomous RNA Silencing
To be Published
|
|
6CV0
 
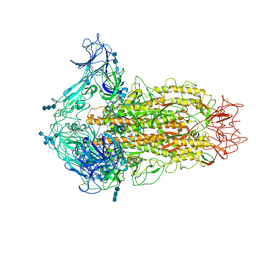 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
7KM5
 
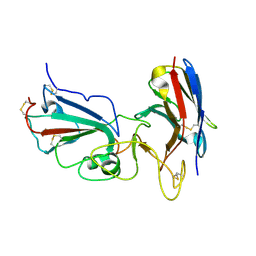 | | Crystal structure of SARS-CoV-2 RBD complexed with Nanosota-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Spike protein S1, ... | | Authors: | Ye, G, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The development of Nanosota - 1 as anti-SARS-CoV-2 nanobody drug candidates.
Elife, 10, 2021
|
|
8JZW
 
 | |
1R8A
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | MANGANESE (II) ION, SODIUM ION, tRNA nucleotidyltransferase | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
2N7J
 
 | |
2LS3
 
 | |
8E7X
 
 | | RsTSPO A138F with one Heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7Y
 
 | | RsTSPO A138F with two heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7Z
 
 | | RsTSPO mutant -A138F | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7W
 
 | | RsTSPO A139T with Heme | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
4YRB
 
 | | mouse TDH mutant R180K with NAD+ bound | | Descriptor: | L-threonine 3-dehydrogenase, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
1M4M
 
 | | Mouse Survivin | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, ZINC ION | | Authors: | Muchmore, S.W, Chen, J, Jakob, C, Zakula, D, Matayoshi, E.D, Wu, W, Zhang, H, Li, F, Ng, S.C, Altieri, D.C. | | Deposit date: | 2002-07-03 | | Release date: | 2002-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE AND MUTAGENIC ANALYSIS OF THE INHIBITOR-OF-APOPTOSIS PROTEIN SURVIVIN
MOL.CELL, 6, 2000
|
|
8JM1
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde from the cyanohydrin cleavage | | Descriptor: | (R)-mandelonitrile lyase, 1,2-ETHANEDIOL, 2,2-dimethyl-4H-1,3-benzodioxine-6-carbaldehyde, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde from the cyanohydrin cleavage
To Be Published
|
|
4YR9
 
 | | mouse TDH with NAD+ bound | | Descriptor: | GLYCEROL, L-threonine 3-dehydrogenase, mitochondrial, ... | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
8GY6
 
 | |
4YRA
 
 | | mouse TDH in the apo form | | Descriptor: | L-threonine 3-dehydrogenase, mitochondrial | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
7VLA
 
 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
