3RZH
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*CP*TP*GP*TP*CP*TP*(ME6)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZG
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*CP*TP*GP*TP*CP*AP*TP*CP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*TP*GP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZM
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*AP*TP*GP*TP*AP*TP*AP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*TP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZK
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(EDA)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*TP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZJ
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(ME6)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
6PB0
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6PB1
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
2B1I
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
2VN3
 
 | | Nitrite Reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
5WYB
 
 | | Structure of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
2VMJ
 
 | | Type 1 Copper-Binding Loop of Nitrite Reductase mutant: 130- CAPEGMVPWHVVSGM-144 to 130-CTPHPFM-136 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2B
 
 | | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2C
 
 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2A
 
 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
3RZL
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*AP*TP*GP*TP*AP*TP*CP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*IP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
1EER
 
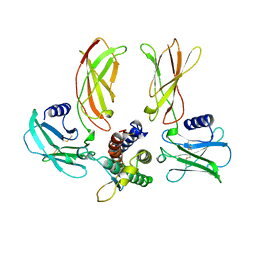 | |
2YJY
 
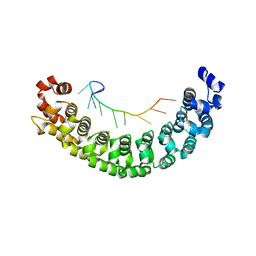 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
5CP3
 
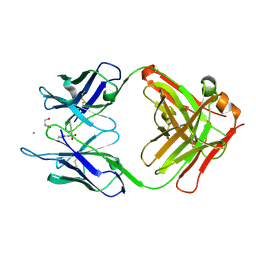 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides in Complex with Sulfathiazole | | Descriptor: | 4-amino-N-(1,3-thiazol-2-yl)benzenesulfonamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
5WYD
 
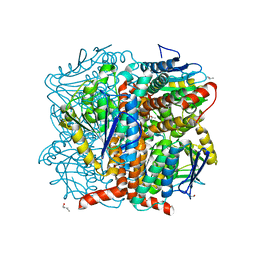 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5J7A
 
 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
