7Z13
 
 | | S. cerevisiae CMGE dimer nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
7QHS
 
 | | S. cerevisiae CMGE nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
4CB7
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8e) | | Descriptor: | 5-[2-(2-azanyl-7-methyl-6-oxidanylidene-1H-purin-9-ium-9-yl)ethanoyl]-2-oxidanyl-benzoic acid, GLYCEROL, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB5
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8f) | | Descriptor: | 9-N-(3-CARBOXY-4-HYDROXYPHENYL)KETOMETHYL-7-N-METHYLGUANINE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB6
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 11) | | Descriptor: | 2,9-N,N-DI(4-CARBOXYBUTYL)-7-N-METHYLGUANINE, IODIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB4
 
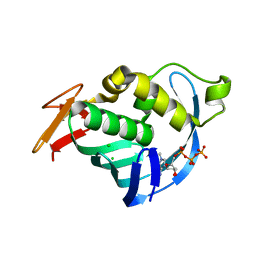 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
2LZG
 
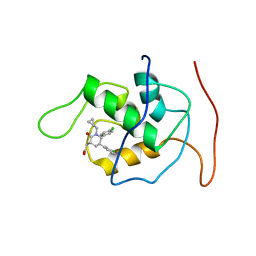 | | NMR Structure of Mdm2 (6-125) with Pip-1 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-(cyclopropylmethyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Michelsen, K.B, Jordan, J.B, Lewis, J, Long, A.M, Yang, E, Rew, Y, Zhou, J, Yakowec, P, Schnier, P.D, Huang, X, Poppe, L. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ordering of the N-Terminus of Human MDM2 by Small Molecule Inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
1BE9
 
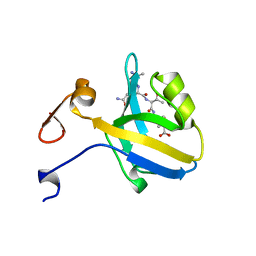 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
2MHS
 
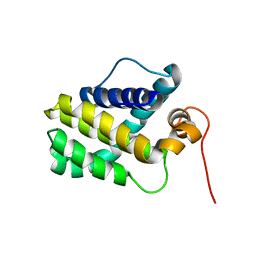 | | NMR Structure of human Mcl-1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, G, Poppe, L, Aoki, K, Yamane, H, Lewis, J, Szyperski, T. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Quality NMR Structure of Human Anti-Apoptotic Protein Domain Mcl-1(171-327) for Cancer Drug Design.
Plos One, 9, 2014
|
|
1BFE
 
 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
2VQZ
 
 | | Structure of the cap-binding domain of influenza virus polymerase subunit PB2 with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Tarendeau, F, Resa-Infante, P, Coloma, R, Crepin, T, Sehr, P, Lewis, J, Ruigrok, R.W.H, Ortin, J, Hart, D.J, Cusack, S. | | Deposit date: | 2008-03-21 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for CAP Binding by Influenza Virus Polymerase Subunit Pb2.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5T88
 
 | | Prolyl oligopeptidase from Pyrococcus furiosus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, PROLINE, ... | | Authors: | Ellis-Guardiola, K, Lewis, J, Sukumar, N. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure and Conformational Dynamics of Pyrococcus furiosus Prolyl Oligopeptidase.
Biochemistry, 58, 2019
|
|
