3LOJ
 
 | | Structure of Mycobacterium tuberculosis dUTPase H145A mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Pecsi, I, Lopata, A, Vertessy, B.G, Toth, J. | | Deposit date: | 2010-02-04 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase
Nucleic Acids Res., 38, 2010
|
|
4GV8
 
 | | DUTPase from phage phi11 of S.aureus: visualization of the species-specific insert | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Leveles, I, Harmat, V, Nemeth, V, Bendes, A, Szabo, J, Kadar, V, Zagyva, I, Rona, G, Toth, J, Vertessy, B.G. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and enzymatic mechanism of a moonlighting dUTPase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3HZA
 
 | | Crystal structure of dUTPase H145W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Pecsi, I, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-06-23 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase.
Nucleic Acids Res., 38, 2010
|
|
3H6D
 
 | | Structure of the mycobacterium tuberculosis DUTPase D28N mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Nagy, G, Takacs, E, Lopata, A, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-04-23 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct contacts between conserved motifs of different subunits provide major contribution to active site organization in human and mycobacterial dUTPases.
Febs Lett., 584, 2010
|
|
3I93
 
 | | Crystal structure of Mycobacterium tuberculosis dUTPase STOP138T mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Lopata, A, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-07-10 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct contacts between conserved motifs of different subunits provide major contribution to active site organization in human and mycobacterial dUTPases.
Febs Lett., 584, 2010
|
|
4WRK
 
 | |
8CGA
 
 | | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, Z.S, Benedek, A, Leveles, I, Vertessy, B.G. | | Deposit date: | 2023-02-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant
To Be Published
|
|
6HDE
 
 | | Structure of Escherichia coli dUTPase Q93H mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Benedek, A, Vertessy, B.G, Leveles, I. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition.
Biomolecules, 9, 2019
|
|
6F6T
 
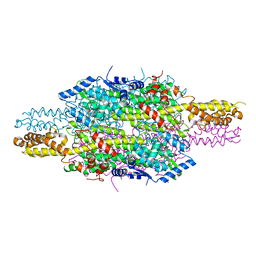 | |
5ECT
 
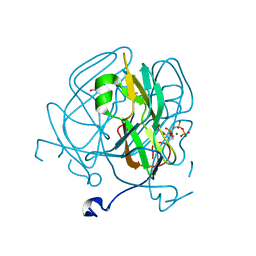 | | Mycobacterium tuberculosis dUTPase G143STOP mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Nagy, G.N, Leveles, I, Harmat, V, Vertessy, G.B. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Characterization of Arginine Fingers: Identification of an Arginine Finger for the Pyrophosphatase dUTPases.
J. Am. Chem. Soc., 138, 2016
|
|
6H2O
 
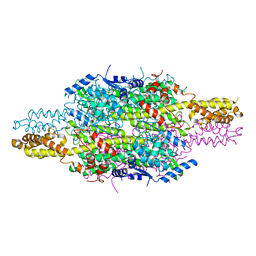 | | APO structure of Phenylalanine ammonia-lyase from Petroselinum crispum | | Descriptor: | Phenylalanine ammonia-lyase 1 | | Authors: | Molnar, B, Bata, Z, Leveles, I, Poppe, L, Vertessy, G.B. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Tunnel Engineering Aided by X-ray Crystallography and Functional Dynamics Swaps the Function of MIO-Enzymes
Acs Catalysis, 2021
|
|
5EDD
 
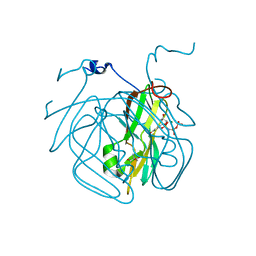 | | Crystal structure of Mycobacterium tuberculosis dUTPase R140K, H145W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Nagy, G.N, Leveles, I, Lopata, A, Harmat, V, Toth, J, Vertessy, G.B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of Arginine Fingers: Identification of an Arginine Finger for the Pyrophosphatase dUTPases.
J. Am. Chem. Soc., 138, 2016
|
|
6HQF
 
 | | Structure of Phenylalanine ammonia-lyase from Petroselinum crispum in complex with (R)-APEP | | Descriptor: | Phenylalanine ammonia-lyase 1, [(1R)-1-amino-2-phenylethyl]phosphonic acid | | Authors: | Bata, Z, Molnar, B, Leveles, I, Poppe, L, Vertessy, G.B. | | Deposit date: | 2018-09-24 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Substrate Tunnel Engineering Aided by X-ray Crystallography and Functional Dynamics Swaps the Function of MIO-Enzymes
Acs Catalysis, 2021
|
|
4GCY
 
 | | Structure of Mycobacterium tuberculosis dUTPase H21W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, J, Vertessy, B.G, Leveles, I, Bendes, A. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RAMD identification of substrate binding pathways to the active site of dUTPase
To be Published
|
|
5NP3
 
 | | Abl2 SH3 | | Descriptor: | Abelson tyrosine-protein kinase 2 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5NP5
 
 | | Abl2 SH3 pTyr116/161 | | Descriptor: | Abelson tyrosine-protein kinase 2, SULFATE ION | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5NP2
 
 | | Abl1 SH3 pTyr89/134 | | Descriptor: | Tyrosine-protein kinase ABL1 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
