7Y8J
 
 | | 3D1 in complex with 6-mer HR1 peptide from SARS-CoV-2 | | Descriptor: | Spike protein S2', heavy chain of 3D1, light chain of 3D1 | | Authors: | Lei, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Cross-reactive epitopes between HIV and Coronavirus revealed by 3D1
To Be Published
|
|
8HQ4
 
 | | B27 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-fluorophenyl)carbonylamino]-4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Lei, Y, Sun, Q. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | B27 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
8TOO
 
 | | Crystal structure of Epstein-Barr virus gp42 in complex with antibody 4C12 | | Descriptor: | 4C12 heavy chain, 4C12 light chain, Glycoprotein 42 | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNN
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
6LQ9
 
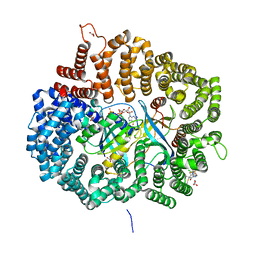 | | S109 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (3~{R},4~{S})-1-[[6-chloranyl-5-(trifluoromethyl)pyridin-2-yl]amino]-3,4-dimethyl-pyrrolidine-2,5-dione, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1.
J.Med.Chem., 64, 2021
|
|
6M6X
 
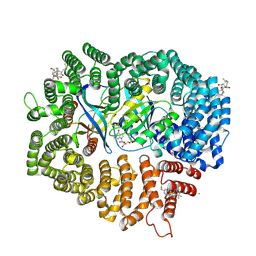 | | Oridonin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
6M60
 
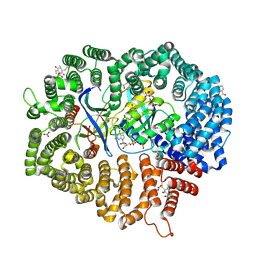 | | Plumbagin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (2~{R})-2-methyl-5-oxidanyl-2,3-dihydronaphthalene-1,4-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
7YPZ
 
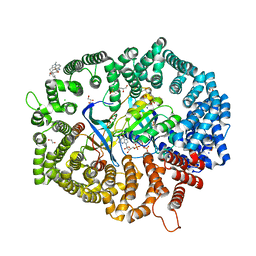 | | Zafirlukast in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Searching for Novel Noncovalent Nuclear Export Inhibitors through a Drug Repurposing Approach.
J.Med.Chem., 66, 2023
|
|
6KYB
 
 | | Crystal structure of Atg18 from Saccharomyces cerevisiae | | Descriptor: | Autophagy-related protein 18 | | Authors: | Tang, D, Lei, Y, Liao, G, Chen, Q, Xu, L, Lu, K, Qi, S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of Atg18 reveals a new binding site for Atg2 in Saccharomyces cerevisiae.
Cell.Mol.Life Sci., 78, 2021
|
|
7CND
 
 | | NCI-1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1.
J.Med.Chem., 64, 2021
|
|
7C9J
 
 | | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Protein-glutamine gamma-glutamyltransferase | | Authors: | Takita, T, Mikami, B, Lei, Y, Jing, Y, Yamada, A, Yasukawa, K. | | Deposit date: | 2020-06-06 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension)
To be published
|
|
5ZPU
 
 | | LFS829 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (Z)-{[(3E)-4-{(R)-[3,5-bis(trifluoromethyl)phenyl]sulfinyl}but-3-en-1-yl]imino}methanethiol, CHLORIDE ION, Exportin-1,Exportin-1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | To be published
To Be Published
|
|
5YWO
 
 | | Structure of JEV-2F2 Fab complex | | Descriptor: | 2F2 heavy chain, 2F2 light chain, JEV E protein, ... | | Authors: | Qiu, X, Lei, Y.F, Yang, P, Gao, Q, Wang, N, Cao, L, Yuan, S, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5YWP
 
 | | JEV-2H4 Fab complex | | Descriptor: | 2H4 heavy chain, 2H4 light chain, JEV E protein, ... | | Authors: | Qiu, X.D, Lei, Y.F, Yang, P, Gao, Q, WANG, N, Cao, L, Yuan, S, Wang, X.X, Xu, Z.K, Rao, Z.H. | | Deposit date: | 2017-11-29 | | Release date: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
7DBG
 
 | | Yeast CRM1e (apo) in complex with Ran-RanBP1 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-10-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
8HQ5
 
 | | G6 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-bromophenyl)carbonylamino]-4-[4-(5-chloranyl-2-methyl-phenyl)piperazin-1-yl]benzoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | F1 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
8HUG
 
 | | F1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 4-[4-(3-chlorophenyl)piperazin-1-yl]-3-[(3-fluorophenyl)sulfonylamino]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUF
 
 | | B28 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-chlorophenyl)carbonylamino]-4-[4-(2,5-dimethylphenyl)piperazin-1-yl]benzoic acid, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
5YWF
 
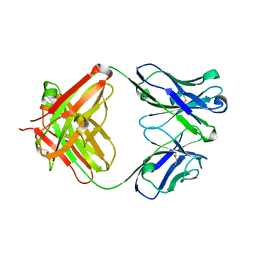 | | Crystal structure of 2H4 Fab | | Descriptor: | 2H4 heavy chain, 2H4 light chain | | Authors: | Qiu, X, Lei, Y, Yang, P, Gao, Q, Wang, N, Cao, L, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
6JTO
 
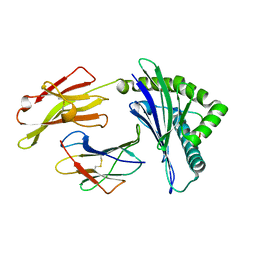 | | Crystal structure of HLA-C05 in complex with a tumor mut10m peptide | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Lei, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational discovery of a cancer neoepitope harboring the KRAS G12D driver mutation.
Sci China Life Sci, 2021
|
|
6JTP
 
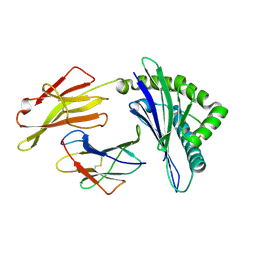 | | Crystal structure of HLA-C08 in complex with a tumor mut9m peptide | | Descriptor: | 9-mer peptide, Beta-2-microglobulin, HLA class I antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Lei, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
7DKM
 
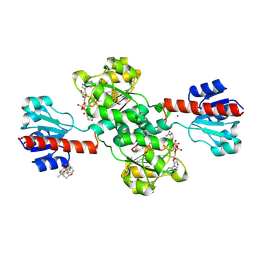 | | PHGDH covalently linked to oridonin | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, CHLORIDE ION, D-3-phosphoglycerate dehydrogenase, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biophysical and biochemical properties of PHGDH revealed by studies on PHGDH inhibitors.
Cell.Mol.Life Sci., 79, 2021
|
|
