2VZ9
 
 | |
2VZ8
 
 | |
1OC6
 
 | | structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
2JSB
 
 | | Solution structure of arenicin-1 | | Descriptor: | Arenicin-1 | | Authors: | Jakovkin, I.B, Hecht, O, Gelhaus, C, Krasnosdembskaya, A.D, Fedders, H, Leippe, M, Groetzinger, J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-05 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of the antimicrobial peptide arenicin
Biochem.J., 410, 2008
|
|
1HF6
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE | | Descriptor: | ACETIC ACID, ENDOGLUCANASE B, GLYCEROL, ... | | Authors: | Varrot, A, Withers, S, Vasella, A, Schulein, M, Davies, G.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2JRZ
 
 | | Solution structure of the Bright/ARID domain from the human JARID1C protein. | | Descriptor: | Histone demethylase JARID1C | | Authors: | Koehler, C, Bishop, S, Dowler, E.F, Diehl, A, Schmieder, P, Leidert, M, Sundstrom, M, Arrowsmith, C.H, Wiegelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Backbone and sidechain 1H, 13C and 15N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein.
Biomol.Nmr Assign., 2, 2008
|
|
1DJF
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | GLN-ALA-PRO-ALA-TYR-LYS-LYS-ALA-ALA-LYS-LYS-LEU-ALA-GLU-SER | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DNG
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL ACIDIC PEPTIDE | | Descriptor: | HUMAN PLATELET FACTOR 4, SEGMENT 59-73 | | Authors: | montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1QHZ
 
 | |
1QI0
 
 | |
6FAI
 
 | | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Scaiola, A, Pena, C, Weisser, M, Boehringer, D, Leibundgut, M, Klingauf-Nerurkar, P, Gerhardy, S, Panse, V.G, Ban, N. | | Deposit date: | 2017-12-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit.
EMBO J., 37, 2018
|
|
3E4A
 
 | | Human IDE-inhibitor complex at 2.6 angstrom resolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, HYDROXAMATE PEPTIDE II1, ... | | Authors: | Malito, E, Leissring, M.A, Choi, S, Cuny, G.D, Tang, W.J. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin.
Plos One, 5, 2010
|
|
7ASD
 
 | | Structure of native royal jelly filaments | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mattei, S, Ban, A, Picenoni, A, Leibundgut, M, Glockshuber, R, Boehringer, D. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of native glycolipoprotein filaments in honeybee royal jelly.
Nat Commun, 11, 2020
|
|
7AVE
 
 | | Perdeuterated refolded hen egg-white lysozyme at 100 K | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Larsen, S, Mossou, E, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
5NFE
 
 | | Neutron structure of human transthyretin (TTR) T119M mutant at room temperature to 1.85A resolution | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Ostermann, A, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.853 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
5NFW
 
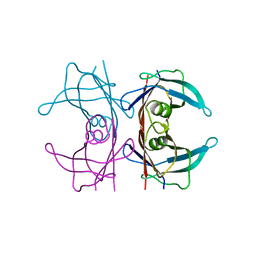 | | Neutron structure of human transthyretin (TTR) S52P mutant at room temperature to 1.8A resolution (quasi-Laue) | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
3F9V
 
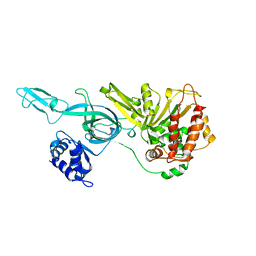 | | Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Chen, X.J, Brewster, A.S, Wang, G.G, Yu, X, Greenleaf, W, Tjajadi, M, Klein, M. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6ZOL
 
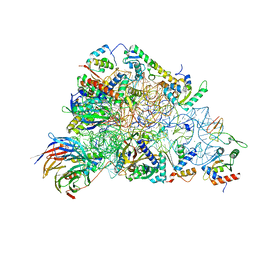 | | SARS-CoV-2-Nsp1-40S complex, focused on head | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOJ
 
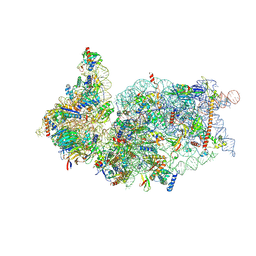 | | SARS-CoV-2-Nsp1-40S complex, composite map | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3F6E
 
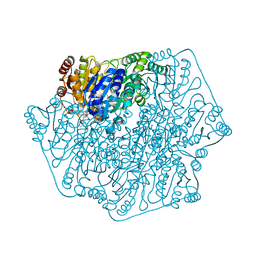 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor 3-PKB | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
3ENG
 
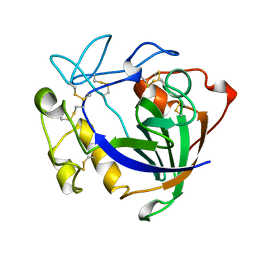 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3F6B
 
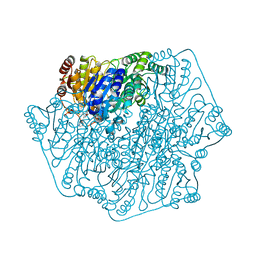 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor PAA | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
3FSJ
 
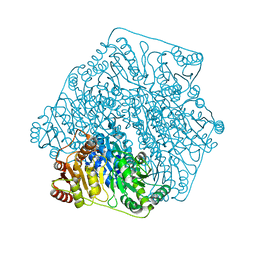 | | Crystal structure of benzoylformate decarboxylase in complex with the inhibitor MBP | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CALCIUM ION | | Authors: | Brandt, G.S, Kenyon, G.L, McLeish, M.J, Jordan, F, Petsko, G.A, Ringe, D. | | Deposit date: | 2009-01-09 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Snapshot of a reaction intermediate: analysis of benzoylformate decarboxylase in complex with a benzoylphosphonate inhibitor.
Biochemistry, 48, 2009
|
|
5LFP
 
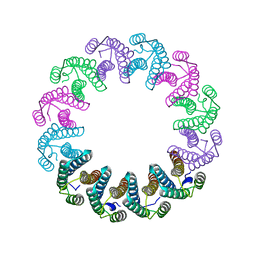 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis (space group P6322, SeMet) | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-23 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
5LZP
 
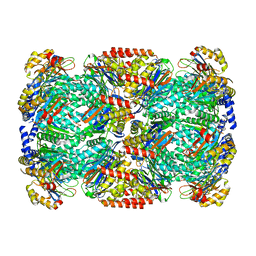 | | Binding of the C-terminal GQYL motif of the bacterial proteasome activator Bpa to the 20S proteasome | | Descriptor: | Bacterial proteasome activator, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
