1Z1Y
 
 | | Crystal structure of Methylated Pvs25, an ookinete protein from Plasmodium vivax | | Descriptor: | YTTERBIUM (III) ION, ookinete surface protein Pvs25 | | Authors: | Saxena, A.K, Singh, K, Su, H.P, Klein, M.M, Stowers, A.W, Saul, A.J, Long, C.A, Garboczi, D.N. | | Deposit date: | 2005-03-07 | | Release date: | 2005-12-06 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The essential mosquito-stage P25 and P28 proteins from Plasmodium form tile-like triangular prisms
Nat.Struct.Mol.Biol., 13, 2006
|
|
1Z3G
 
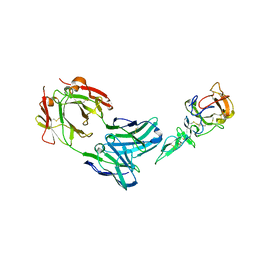 | | Crystal structure of complex between Pvs25 and Fab fragment of malaria transmission blocking antibody 2A8 | | Descriptor: | 2A8 Fab Heavy Chain, 2A8 Fab Light Chain, ookinete surface protein Pvs25 | | Authors: | Saxena, A.K, Singh, K, Su, H.P, Klein, M.M, Stowers, A.W, Saul, A.J, Long, C.A, Garboczi, D.N. | | Deposit date: | 2005-03-12 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The essential mosquito-stage P25 and P28 proteins from Plasmodium form tile-like triangular prisms
Nat.Struct.Mol.Biol., 13, 2006
|
|
4Y36
 
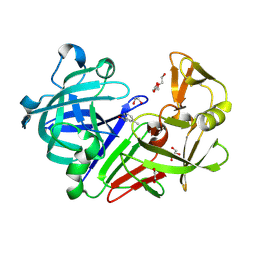 | | Endothiapepsin in complex with fragment 4 | | Descriptor: | 1,2-ETHANEDIOL, 3-methylbenzohydrazide, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
1OJJ
 
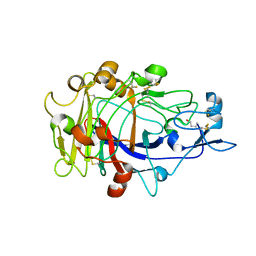 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1N7I
 
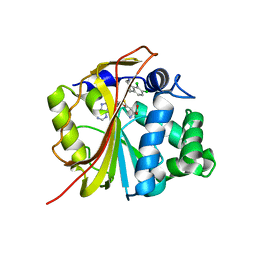 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and the inhibitor LY134046 | | Descriptor: | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO-1H-BENZO[C]AZEPINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1N7J
 
 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and an iodinated inhibitor | | Descriptor: | 7-IODO-1,2,3,4-TETRAHYDRO-ISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
2QKH
 
 | | Crystal structure of the extracellular domain of human GIP receptor in complex with the hormone GIP | | Descriptor: | Cyclic 2,3-di-O-methyl-alpha-D-glucopyranose-(1-4)-2-O-methyl-alpha-D-glucopyranose-(1-4)-2,6-di-O-methyl-alpha-D-glucopyranose-(1-4)-2-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-3-O-methyl-alpha-D-glucopyranose, D(-)-TARTARIC ACID, Glucose-dependent insulinotropic polypeptide, ... | | Authors: | Parthier, C, Kleinschmidt, M, Neumann, P, Rudolph, R, Manhart, S, Schlenzig, D, Fanghanel, J, Rahfeld, J.-U, Demuth, H.-U, Stubbs, M.T. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4OC6
 
 | | Structure of Cathepsin D with inhibitor 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide, Cathepsin D heavy chain, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2LLR
 
 | |
1A39
 
 | | HUMICOLA INSOLENS ENDOCELLULASE EGI S37W, P39W DOUBLE-MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Ducros, V, Lewis, R.J, Borchert, T.V, Schulein, M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oligosaccharide specificity of a family 7 endoglucanase: insertion of potential sugar-binding subsites.
J.Biotechnol., 57, 1997
|
|
2R24
 
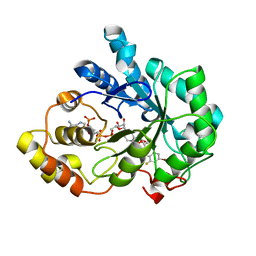 | | Human Aldose Reductase structure | | Descriptor: | Aldose reductase, IDD594, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O.N, Cousido-Siah, A, Haertlein, M, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-24 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.752 Å), X-RAY DIFFRACTION | | Cite: | Quantum model of catalysis based on mobile proton revealed by subatomic X-Ray and neutron diffraction studies of h-Aldose Reductase
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2I59
 
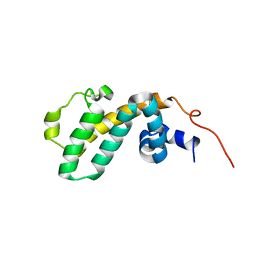 | | Solution structure of RGS10 | | Descriptor: | Regulator of G-protein signaling 10 | | Authors: | Fedorov, O, Higman, V.A, Diehl, A, Leidert, M, Lemak, A, Schmieder, P, Oschkinat, H, Elkins, J, Soundarajan, M, Doyle, D.A, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2HZ8
 
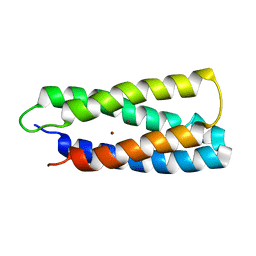 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
4GM1
 
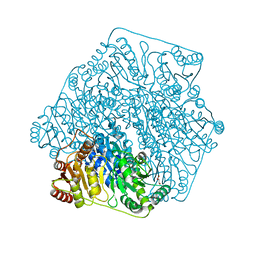 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403S | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
2BVW
 
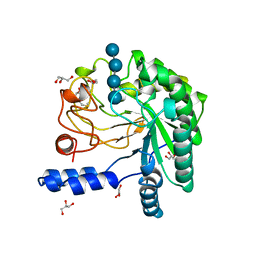 | | CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, GLYCEROL, ... | | Authors: | Varrot, A, Davies, G.J, Schulein, M. | | Deposit date: | 1999-02-18 | | Release date: | 2000-02-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding.
Biochemistry, 38, 1999
|
|
2CKY
 
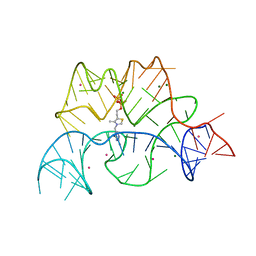 | | Structure of the Arabidopsis thaliana thiamine pyrophosphate riboswitch with its regulatory ligand | | Descriptor: | MAGNESIUM ION, NUCLEIC ACID, OSMIUM ION, ... | | Authors: | Thore, S, Leibundgut, M, Ban, N. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with its Regulatory Ligand.
Science, 312, 2006
|
|
2CDH
 
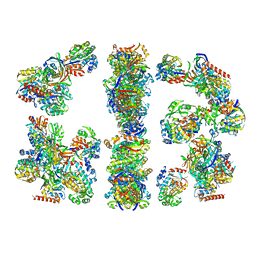 | | ARCHITECTURE OF THE THERMOMYCES LANUGINOSUS FUNGAL FATTY ACID SYNTHASE AT 5 ANGSTROM RESOLUTION. | | Descriptor: | DEHYDRATASE, ENOYL REDUCTASE, KETOACYL REDUCTASE, ... | | Authors: | Jenni, S, Leibundgut, M, Maier, T, Ban, N. | | Deposit date: | 2006-01-24 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Architecture of a Fungal Fatty Acid Synthase at 5 A Resolution.
Science, 311, 2006
|
|
4GG1
 
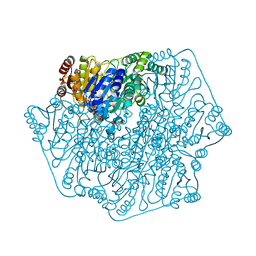 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403T | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-04 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
4K9O
 
 | |
2H1L
 
 | |
1DYS
 
 | | Endoglucanase CEL6B from Humicola insolens | | Descriptor: | ENDOGLUCANASE | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, M, Varrot, A, Schulein, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Humicola Insolens Family 6 Cellulases: Structure of the Endoglucanase, Cel6B, at 1.6 A Resolution
Biochem.J., 348, 2000
|
|
4K9L
 
 | |
1OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH NON-HYDROLYSABLE SUBSTRATE ANALOGUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-1,4-dithio-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group.
Biochemistry, 35, 1996
|
|
2V2H
 
 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-06-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed-Like Conformation in Dimeric and Monomeric Tim.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5DX6
 
 | |
