1SJG
 
 | | Solution Structure of T4moC, the Rieske Ferredoxin Component of the Toluene 4-Monooxygenase Complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Toluene-4-monooxygenase system protein C | | Authors: | Skjeldal, L, Peterson, F.C, Doreleijers, J.F, Moe, L.A, Pikus, J.D, Volkman, B.F, Westler, W.M, Markley, J.L, Fox, B.G. | | Deposit date: | 2004-03-03 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of T4moC, the Rieske ferredoxin component of the toluene 4-monooxygenase complex
J.Biol.Inorg.Chem., 9, 2004
|
|
3SAI
 
 | |
3SA1
 
 | |
3SA2
 
 | |
3S9U
 
 | |
1DMS
 
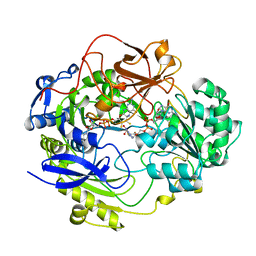 | | STRUCTURE OF DMSO REDUCTASE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM (IV)OXIDE | | Authors: | Schneider, F, Loewe, J, Huber, R, Schindelin, H, Kisker, C, Knaeblein, J. | | Deposit date: | 1996-09-03 | | Release date: | 1998-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 A resolution.
J.Mol.Biol., 263, 1996
|
|
1H70
 
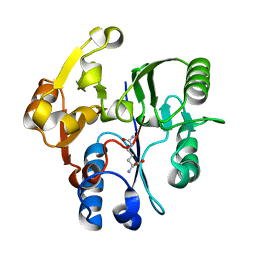 | | DDAH FROM PSEUDOMONAS AERUGINOSA. C249S MUTANT COMPLEXED WITH CITRULLINE | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE | | Authors: | Murray-Rust, J, Leiper, J, McAlister, M, Phelan, J, Tilley, S, Santamaria, J, Vallance, P, McDonald, N. | | Deposit date: | 2001-06-30 | | Release date: | 2001-08-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase.
Nat. Struct. Biol., 8, 2001
|
|
2V3W
 
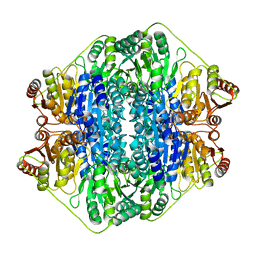 | | Crystal structure of the benzoylformate decarboxylase variant L461A from Pseudomonas putida | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Gocke, D, Walter, L, Gauchenova, K, Kolter, G, Knoll, M, Berthold, C.L, Schneider, G, Pleiss, J, Mueller, M, Pohl, M. | | Deposit date: | 2007-06-25 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Protein Design of Thdp-Dependent Enzymes-Engineering Stereoselectivity.
Chembiochem, 9, 2008
|
|
2JAI
 
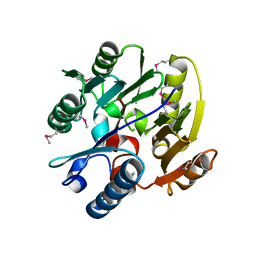 | | DDAH1 complexed with citrulline | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
2JAJ
 
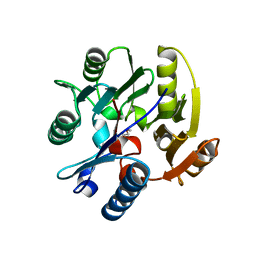 | | DDAH1 complexed with L-257 | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, N~5~-{IMINO[(2-METHOXYETHYL)AMINO]METHYL}-L-ORNITHINE | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
2KGK
 
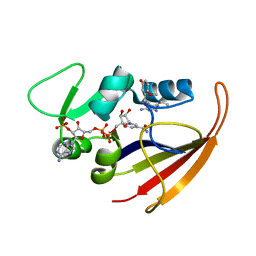 | | Solution structure of Bacillus anthracis dihydrofolate reductase | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Deshmukh, L, Vinogradova, O, Beierlein, J.M, Frey, K.M, Anderson, A.C. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Bacillus anthracis dihydrofolate reductase yields insight into the analysis of structure-activity relationships for novel inhibitors.
Biochemistry, 48, 2009
|
|
8A9Q
 
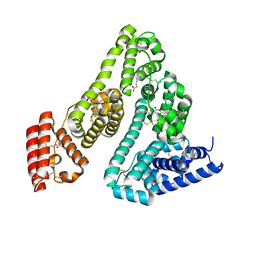 | | Computational design of stable mammalian serum albumins for bacterial expression | | Descriptor: | Albumin, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Khersonsky, O, Dym, O, Fleishman, J.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stable Mammalian Serum Albumins Designed for Bacterial Expression.
J.Mol.Biol., 435, 2023
|
|
6FHE
 
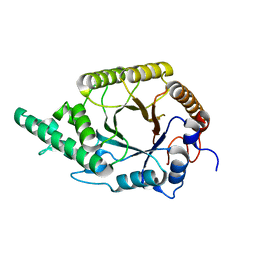 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Synthetic construct | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
5MTI
 
 | | Bamb_5917 Acyl-Carrier Protein | | Descriptor: | Phosphopantetheine-binding protein | | Authors: | Gallo, A, Kosol, S, Griffiths, D, Masschelein, J, Alkhalaf, L, Smith, H, Valentic, T, Tsai, S, Challis, G, Lewandowski, J.R. | | Deposit date: | 2017-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase
Nat.Chem., 2019
|
|
6TBC
 
 | | Crystal structure of S. aureus FabI in complex with NADPH and kalimantacin B | | Descriptor: | (2~{E},5~{R},10~{E},12~{E},15~{S},19~{R})-20-[[(2~{R},3~{R})-3-aminocarbonyloxy-2-methyl-butanoyl]amino]-3,5,15-trimethyl-7-methylidene-19-oxidanyl-17-oxidanylidene-icosa-2,10,12-trienoic acid, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fage, C.D, Masschelein, J. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6TBB
 
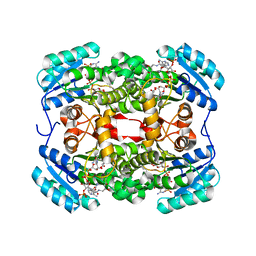 | | Crystal structure of S. aureus FabI in complex with NADPH and kalimantacin A (batumin) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], Kalimantacin, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fage, C.D, Masschelein, J. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6FHF
 
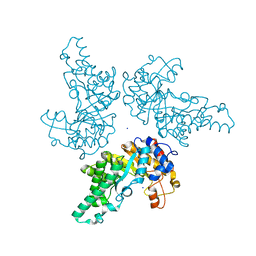 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Design, SODIUM ION | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
3HNA
 
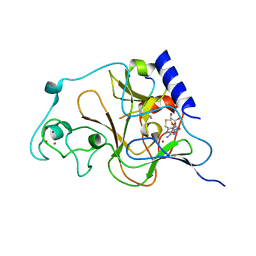 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and mono-Methylated H3K9 Peptide | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, Mono-Methylated H3K9 Peptide, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Wleigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
6T26
 
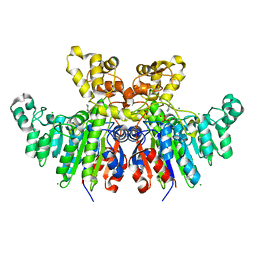 | | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Asgeirsson, B, Hjorleifsson, J.G, Markusson, S, Helland, R. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine.
Biochem Biophys Rep, 24, 2020
|
|
3E0B
 
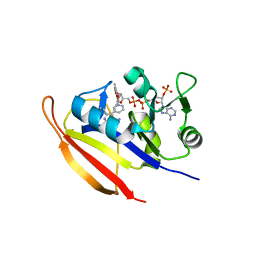 | | Bacillus anthracis Dihydrofolate Reductase complexed with NADPH and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Frey, K.M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthetic and Crystallographic Studies of a New Inhibitor Series Targeting Bacillus anthracis Dihydrofolate Reductase
J.Med.Chem., 51, 2008
|
|
3JWC
 
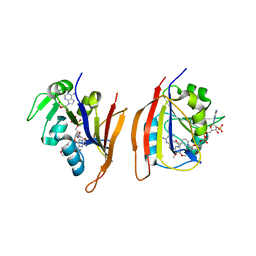 | | Crystal structure of Bacillus anthracis (Y102F) dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-(3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120A) | | Descriptor: | 6-ethyl-5-[3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Mutations in Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships
J.Med.Chem., 2010
|
|
3JWK
 
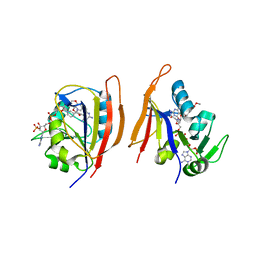 | | Crystal structure of Bacillus anthracis (Y102F) dihydrofolate reductase complexed with NADPH and (S)-2,4-diamino-5-(3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP114A) | | Descriptor: | 5-[(3S)-3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeted Mutations in Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships
J.Med.Chem., 2010
|
|
3JWF
 
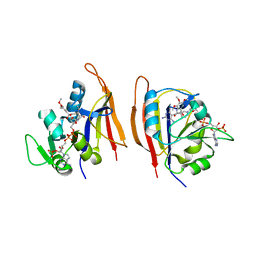 | | Crystal structure of Bacillus anthracis (Y102F) dihydrofolate reductase complexed with NADPH and (R)-2,4-diamino-5-(3-hydroxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP113A) | | Descriptor: | (1R)-3-(2,4-diamino-6-methylpyrimidin-5-yl)-1-(3,4,5-trimethoxyphenyl)prop-2-yn-1-ol, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Mutations in Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships
J.Med.Chem., 2010
|
|
3JVX
 
 | | Crystal structure of Bacillus anthracis dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-(3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120A) | | Descriptor: | 6-ethyl-5-[3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutational Studies into Trimethoprim Resistance in Bacillus anthracis Dihydrofolate Reductase
To be Published
|
|
3JW5
 
 | |
