6TKR
 
 | | Tankyrase 2 in complex with an inhibitor (OM-1704) | | Descriptor: | Tankyrase-2, ZINC ION, ~{N}-[3-[5-(5-ethoxypyridin-2-yl)-4-(2-fluorophenyl)-1,2,4-triazol-3-yl]cyclobutyl]-1,5-naphthyridine-2-carboxamide | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2019-11-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor.
J.Med.Chem., 63, 2020
|
|
7O6X
 
 | | Tankyrase 2 in complex with an inhibitor (OM-153) | | Descriptor: | N-[3-[5-(5-ethoxypyridin-2-yl)-4-(2-fluorophenyl)-1,2,4-triazol-3-yl]cyclobutyl]quinoxaline-5-carboxamide, Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2021-04-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a 1,2,4-Triazole-Based Lead Tankyrase Inhibitor: Part II.
J.Med.Chem., 64, 2021
|
|
3B6E
 
 | | Crystal structure of human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SODIUM ION | | Authors: | Karlberg, T, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain.
To be Published
|
|
7AEO
 
 | | Human ARTD2 in complex with DNA oligonucleotides | | Descriptor: | DNA, Poly [ADP-ribose] polymerase 2, SULFATE ION | | Authors: | Obaji, E, Maksimainen, M.M, Galera-Prat, A, Lehtio, L. | | Deposit date: | 2020-09-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation of PARP2/ARTD2 by DNA damage induces conformational changes relieving enzyme autoinhibition.
Nat Commun, 12, 2021
|
|
1F9C
 
 | | CRYSTAL STRUCTURE OF MLE D178N VARIANT | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE I) | | Authors: | Kajander, T, Lehtio, L, Kahn, P.C, Goldman, A. | | Deposit date: | 2000-07-10 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Buried charged surface in proteins.
Structure Fold.Des., 8, 2000
|
|
4UVU
 
 | | Crystal structure of human tankyrase 2 in complex with 1-((4-(5- methyl-1-oxo-1,2-dihydroisoquinolin-3-yl)phenyl)methyl)pyrrolidin-1- ium | | Descriptor: | 5-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVS
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- pentyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-pentylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UX4
 
 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-(4- methylphenyl)-5-oxo-5,6-dihydro-1,6-naphthyridin-1-ium | | Descriptor: | (1S)-1-methyl-7-(4-methylphenyl)-5-oxo-1,5-dihydro-1,6-naphthyridin-1-ium, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-19 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis and Evaluation in Vitro of Arylnaphthyridinones, Arylpyridopyrimidinones and Their Tetrahydro Derivatives as Inhibitors of the Tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
4UVY
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methoxy-1,2- dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVX
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-fluoro-1,2-dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-fluoroisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVP
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- ethyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-ethylisoquinolin-1(2H)-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
8R79
 
 | | The D2 domain of human DTX3L | | Descriptor: | E3 ubiquitin-protein ligase DTX3L, SULFATE ION | | Authors: | Vela-Rodriguez, C, Lehtio, L, Maksimainen, M, Duman, R, Wagner, A, Glumoff, T. | | Deposit date: | 2023-11-24 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oligomerisation mediated by the D2 domain of DTX3L is critical for DTX3L-PARP9 reading function of mono-ADP-ribosylated androgen receptor.
Biorxiv, 2023
|
|
8TV6
 
 | | SARS-CoV-2 Mac1 in complex with MDOLL-0169 | | Descriptor: | (1R,6R)-6-{[3-(methoxycarbonyl)-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophen-2-yl]carbamoyl}cyclohex-3-ene-1-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of 2-Amide-3-methylester Thiophenes that Target SARS-CoV-2 Mac1 and Repress Coronavirus Replication, Validating Mac1 as an Antiviral Target.
J.Med.Chem., 67, 2024
|
|
8TV7
 
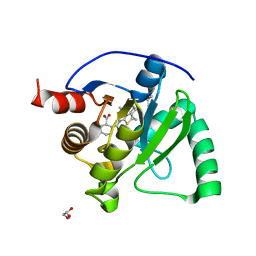 | | SARS-CoV-2 Mac1 in complex with MDOLL-0229 | | Descriptor: | (1R,2R)-2-{[3-(methoxycarbonyl)-4,5,6,7,8,9-hexahydrocycloocta[b]thiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, GLYCEROL, Papain-like protease nsp3 | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-Amide-3-methylester Thiophenes that Target SARS-CoV-2 Mac1 and Repress Coronavirus Replication, Validating Mac1 as an Antiviral Target.
J.Med.Chem., 67, 2024
|
|
7AAH
 
 | | Structure of human pERp1 | | Descriptor: | CITRIC ACID, Marginal zone B- and B1-cell-specific protein | | Authors: | Sowa, S.T, Moilanen, A, Biterova, E, Saaranen, M.J, Lehtio, L, Ruddock, L.W. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution Crystal Structure of Human pERp1, A Saposin-like Protein Involved in IgA, IgM and Integrin Maturation in the Endoplasmic Reticulum.
J.Mol.Biol., 433, 2021
|
|
2ENX
 
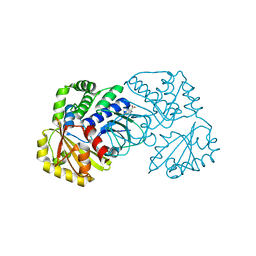 | | Structure of the family II inorganic pyrophosphatase from Streptococcus agalactiae at 2.8 resolution | | Descriptor: | IMIDODIPHOSPHORIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Rantanen, M.K, Lehtio, L, Rajagopal, L, Rubens, C.E, Goldman, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Streptococcus agalactiae family II inorganic pyrophosphatase at 2.80 A resolution
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
6Y73
 
 | | The crystal structure of human MACROD2 in space group P43 | | Descriptor: | ADP-ribose glycohydrolase MACROD2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3BHD
 
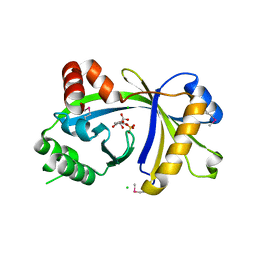 | | Crystal structure of human thiamine triphosphatase (THTPA) | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Busam, R.D, Lehtio, L, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Thiamine Triphosphatase.
To be Published
|
|
7R3Z
 
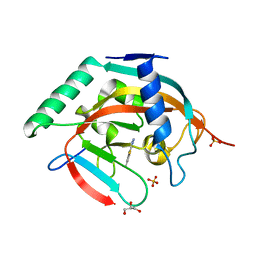 | | Tankyrase 2 catalytic domain in complex with OUL40 | | Descriptor: | 6-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, GLYCEROL, Poly [ADP-ribose] polymerase tankyrase-2, ... | | Authors: | Murthy, S, Venkannagari, H, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7R5X
 
 | | Tankyrase 2 in complex with an inhibitor (OUL211) | | Descriptor: | 7-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, GLYCEROL, Poly [ADP-ribose] polymerase tankyrase-2, ... | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2022-02-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
6Y4Z
 
 | | The crystal structure of human MACROD2 in space group P43212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y4Y
 
 | | The crystal structure of human MACROD2 in space group P41212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3BER
 
 | | Human DEAD-box RNA-helicase DDX47, conserved domain I in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Probable ATP-dependent RNA helicase DDX47, ... | | Authors: | Karlberg, T, Busam, R.D, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3B7G
 
 | | Human DEAD-box RNA helicase DDX20, Conserved domain I (DEAD) in complex with AMPPNP (Adenosine-(Beta,gamma)-imidotriphosphate) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
7Z1Y
 
 | | PARP15 catalytic domain in complex with OUL245 | | Descriptor: | DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15, [1,2,4]triazolo[3,4-b][1,3]benzothiazol-6-ol | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
