1QKG
 
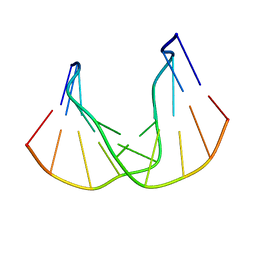 | | DNA DECAMER DUPLEX CONTAINING T-T DEWAR PHOTOPRODUCT | | 分子名称: | DNA (5'-D(*CP*GP*CP*AP*(HYD)TP*+TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | 著者 | Lee, J.-H, Bae, S.-H, Choi, Y.-J, Choi, B.-S. | | 登録日 | 1999-07-20 | | 公開日 | 2000-05-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dewar Photoproduct of Thymidylyl(3'-->5')-Thymidine (Dewar Product) Exhibits Mutagenic Behavior in Accordance with the "A Rule".
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7JP1
 
 | |
5GPC
 
 | |
7JOY
 
 | |
5FUU
 
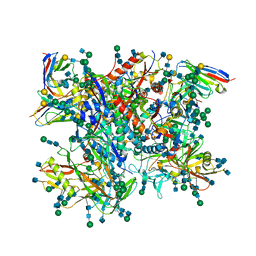 | | Ectodomain of cleaved wild type JR-FL EnvdCT trimer in complex with PGT151 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lee, J.H, Ward, A.B. | | 登録日 | 2016-01-29 | | 公開日 | 2016-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-Em Structure of a Native, Fully Glycosylated and Cleaved HIV-1 Envelope Trimer
Science, 351, 2016
|
|
7KHP
 
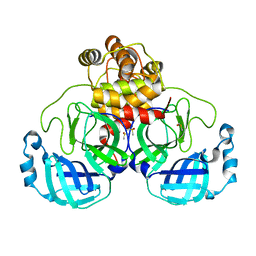 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2020-10-21 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
1D5R
 
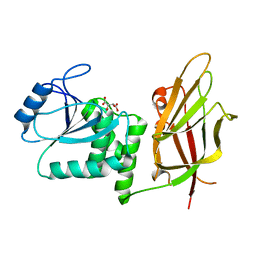 | | Crystal Structure of the PTEN Tumor Suppressor | | 分子名称: | L(+)-TARTARIC ACID, PHOSPHOINOSITIDE PHOSPHATASE PTEN | | 著者 | Lee, J.O, Yang, H, Georgescu, M.-M, Di Cristofano, A, Pavletich, N.P. | | 登録日 | 1999-10-11 | | 公開日 | 1999-11-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association.
Cell(Cambridge,Mass.), 99, 1999
|
|
4MVC
 
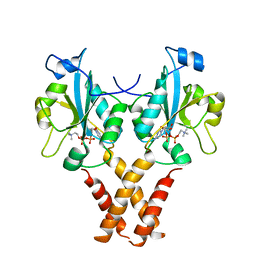 | | Crystal Structure of a Mammalian Cytidylyltransferase | | 分子名称: | Choline-phosphate cytidylyltransferase A, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | 著者 | Lee, J, Cornell, R.B. | | 登録日 | 2013-09-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis for Autoinhibition of CTP:Phosphocholine Cytidylyltransferase (CCT), the Regulatory Enzyme in Phosphatidylcholine Synthesis, by Its Membrane-binding Amphipathic Helix.
J.Biol.Chem., 289, 2014
|
|
2F2B
 
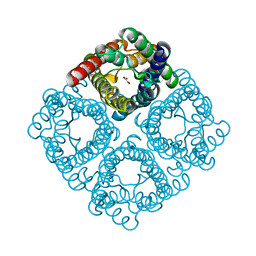 | | Crystal structure of integral membrane protein Aquaporin AqpM at 1.68A resolution | | 分子名称: | Aquaporin aqpM, GLYCEROL | | 著者 | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2005-11-15 | | 公開日 | 2005-12-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3C4T
 
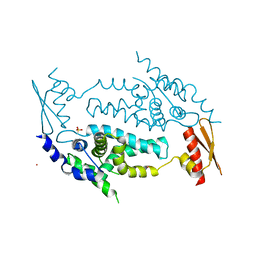 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | 分子名称: | CADMIUM ION, Endoribonuclease Dicer | | 著者 | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | 登録日 | 2008-01-30 | | 公開日 | 2008-02-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | 分子名称: | Candidapepsin-5, Pepstatin | | 著者 | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | 登録日 | 2007-08-17 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
4Q16
 
 | |
7DOG
 
 | |
3PD7
 
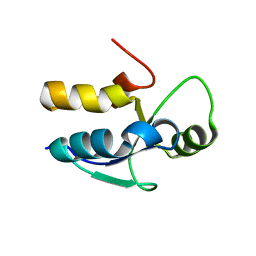 | |
3N8X
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ... | | 著者 | Lee, J.Y. | | 登録日 | 2010-05-28 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
3C4B
 
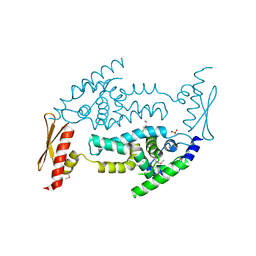 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | 分子名称: | Endoribonuclease Dicer | | 著者 | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | 登録日 | 2008-01-29 | | 公開日 | 2008-02-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3EQA
 
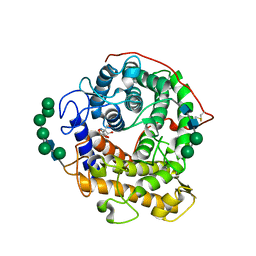 | | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glucoamylase, ... | | 著者 | Lee, J, Paetzel, M. | | 登録日 | 2008-09-30 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the catalytic domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3O4A
 
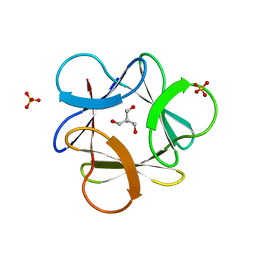 | |
7CV2
 
 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | 分子名称: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | 著者 | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | 登録日 | 2020-08-25 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
1GUX
 
 | |
3Q8W
 
 | | A b-aminoacyl containing thiazolidine derivative and DPPIV complex | | 分子名称: | Dipeptidyl peptidase 4, N-(4-{[({(2R)-3-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]-1,3-thiazolidin-2-yl}carbonyl)amino]methyl}phenyl)-D-valine | | 著者 | Lee, J.O, Song, D.H. | | 登録日 | 2011-01-07 | | 公開日 | 2011-03-16 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Discovery of b-aminoacyl containing thiazolidine derivatives as potent and selective dipeptidyl peptidase IV inhibitors
To be Published
|
|
7CV0
 
 | | Crystal structure of B. halodurans NiaR in apo form | | 分子名称: | Transcriptional regulator NiaR, ZINC ION | | 著者 | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | 登録日 | 2020-08-25 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | 分子名称: | ADENINE, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2001-09-13 | | 公開日 | 2002-10-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
1Z2V
 
 | |
3OL0
 
 | |
