5I2O
 
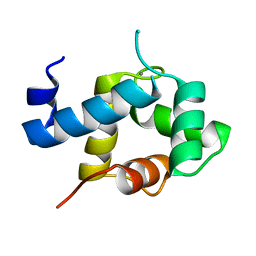 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
4L1C
 
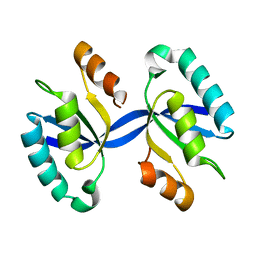 | | Crystal structure of Dimerized N-terminal Domain of MinC | | Descriptor: | Probable septum site-determining protein MinC | | Authors: | An, J.Y, Kim, T.G, Park, K.R, Lee, J.G, Youn, H.S, Kang, J.Y, Lee, Y, Kang, G.B, Eom, S.H. | | Deposit date: | 2013-06-03 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the N-terminal domain of MinC dimerized via domain swapping.
J Synchrotron Radiat, 20, 2013
|
|
4MHQ
 
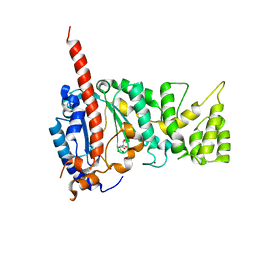 | | Crystal structure of human primase catalytic subunit | | Descriptor: | CITRIC ACID, DNA primase small subunit, ZINC ION | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human primase catalytic subunit
To be Published
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
2RSY
 
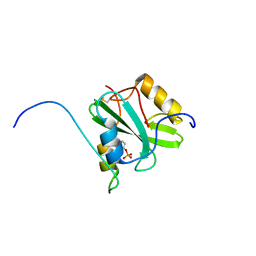 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
4I9A
 
 | | Crystal Structure of Sus scrofa Quinolinate Phosphoribosyltransferase in Complex with Nicotinate Mononucleotide | | Descriptor: | NICOTINATE MONONUCLEOTIDE, quinolinate phosphoribosyltransferase | | Authors: | Youn, H.-S, Kim, M.-K, Kang, K.B, Kim, T.G, Lee, J.-G, An, J.Y, Park, K.R, Lee, Y, Kang, J.Y, Song, H.E, Park, I, Cho, C, Fukuoka, S, Eom, S.H. | | Deposit date: | 2012-12-05 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Crystal structure of Sus scrofa quinolinate phosphoribosyltransferase in complex with nicotinate mononucleotide
Plos One, 8, 2013
|
|
5ZCU
 
 | | Crystal structure of RCAR3:PP2C wild-type with pyrabactin | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, Y, Lee, S. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural determinants for pyrabactin recognition in ABA receptors in Oryza sativa.
Plant Mol.Biol., 100, 2019
|
|
5H0P
 
 | | Crystal structure of EF-hand protein mutant | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, An, J.Y, Kang, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Mun, S.A, Jun, C.D, Song, W.K, Eom, S.H. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural mechanism underlying regulation of human EFhd2/Swiprosin-1 actin-bundling activity by Ser183 phosphorylation.
Biochem. Biophys. Res. Commun., 483, 2017
|
|
4MM2
 
 | | Crystal structure of yeast primase catalytic subunit | | Descriptor: | CADMIUM ION, CITRIC ACID, DNA primase small subunit | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-09-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast primase catalytic subunit
To be Published
|
|
8JUU
 
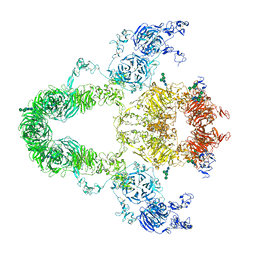 | | rat megalin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JX8
 
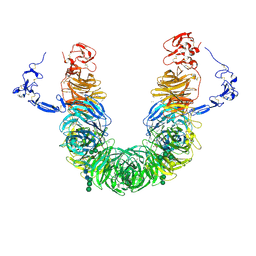 | | rat megalin head | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JX9
 
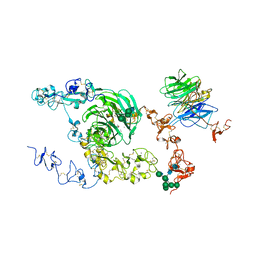 | | rat megalin bodyA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXH
 
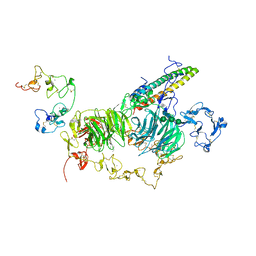 | | rat megalin RAP complex wingA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXF
 
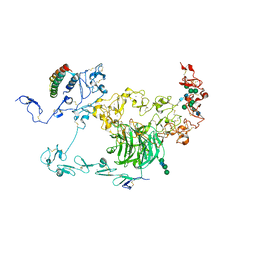 | | rat megalin RAP complex bodyA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin receptor-associated protein, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXJ
 
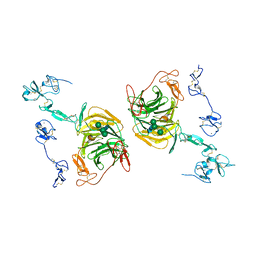 | | rat megalin RAP complex leg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LDL receptor related protein 2, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXD
 
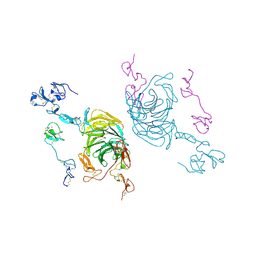 | | Cryo-EM structure of rat megalin leg | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JUT
 
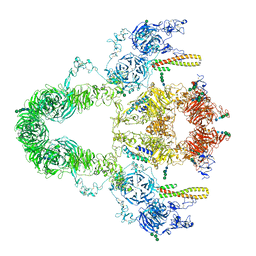 | | rat megalin RAP complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXE
 
 | | rat megalin RAP complex head | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXI
 
 | | rat megalin RAP complex wingB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXC
 
 | | rat megalin wingB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXG
 
 | | rat megalin RAP complex bodyB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXA
 
 | | cryo-EM structure of rat megalin bodyB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXB
 
 | | Cryo-EM structure of rat megalin wingAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2RTT
 
 | | Solution structure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor | | Descriptor: | ChiC | | Authors: | Okumura, A, Uemura, M, Yamada, N, Chikaishi, E, Takai, T, Yoshio, S, Akagi, K, Morita, J, Lee, Y, Yokogawa, D, Suzuki, K, Watanabe, T, Ikegami, T. | | Deposit date: | 2013-08-26 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chitin-binding domain of chitinase Chi18aC from Streptomyces coelicolor
To be Published
|
|
