1IQO
 
 | | Solution structure of MTH1880 from methanobacterium thermoautotrophicum | | Descriptor: | HYPOTHETICAL PROTEIN MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-23 | | Release date: | 2002-07-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
1RJA
 
 | |
1IQS
 
 | | Minimized average structure of MTH1880 from Methanobacterium Thermoautotrophicum | | Descriptor: | MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-29 | | Release date: | 2002-07-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
8HU2
 
 | | Rattus Syntenin-1 PDZ domain with inhibitor | | Descriptor: | (2~{S})-2-(9~{H}-fluoren-9-ylmethoxycarbonylamino)-3-(4-oxidanylidene-5~{H}-pyrimidin-2-yl)propanoic acid, Syntenin-1 | | Authors: | Heo, Y, Lee, J, Yun, J.H, Lee, W. | | Deposit date: | 2022-12-22 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of STNPDZ with inhibitor at 1.60 Angstroms resolution.
To Be Published
|
|
6WQE
 
 | | Solution Structure of the IWP-051-bound H-NOX from Shewanella woodyi in the Fe(II)CO ligation state | | Descriptor: | 5-fluoro-2-{1-[(2-fluorophenyl)methyl]-5-(1,2-oxazol-3-yl)-1H-pyrazol-3-yl}pyrimidin-4-ol, CARBON MONOXIDE, Heme NO binding domain protein, ... | | Authors: | Chen, C.Y, Lee, W, Montfort, W.R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051.
Protein Sci., 30, 2021
|
|
1EJP
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN | | Descriptor: | SYNDECAN-4 | | Authors: | Lee, D, Oh, E.S, Woods, A, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-03 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric cytoplasmic domain of syndecan-4.
Biochemistry, 40, 2001
|
|
6OCV
 
 | |
3W8L
 
 | | Crystal structure of human CK2 in complex with inositol hexakisphosphate | | Descriptor: | Casein kinase II subunit alpha, INOSITOL HEXAKISPHOSPHATE | | Authors: | Son, S.H, Lee, W.-K, Yu, Y.G, Lee, H.H. | | Deposit date: | 2013-03-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
6CGH
 
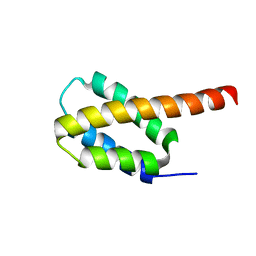 | | Solution structure of the four-helix bundle region of human J-protein Zuotin, a component of ribosome-associated complex (RAC) | | Descriptor: | DnaJ homolog subfamily C member 2 | | Authors: | Shrestha, O.K, Lee, W, Tonelli, M, Cornilescu, G, Markley, J.L, Ciesielski, S.J, Craig, E.A. | | Deposit date: | 2018-02-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and evolution of the 4-helix bundle domain of Zuotin, a J-domain protein co-chaperone of Hsp70.
Plos One, 14, 2019
|
|
2NLU
 
 | |
2KCC
 
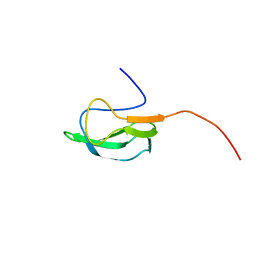 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
3TD5
 
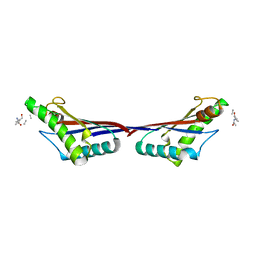 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD3
 
 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | Descriptor: | GLYCINE, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
2GKI
 
 | | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity | | Descriptor: | nuclease | | Authors: | Kim, Y.R, Kim, J.S, Lee, S.H, Lee, W.R, Sohn, J.N, Chung, Y.C, Shim, H.K, Lee, S.C, Kwon, M.H, Kim, Y.S. | | Deposit date: | 2006-04-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity.
J.Biol.Chem., 281, 2006
|
|
6LVU
 
 | |
8HNJ
 
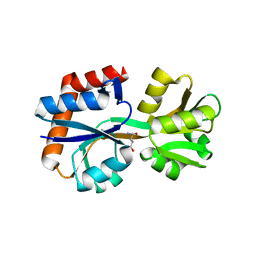 | | Domain-stabilized glutamine-binding protein | | Descriptor: | GLUTAMINE, Glutamine ABC transporter, periplasmic glutamine-binding protein GlnH | | Authors: | Choi, S.H, Park, J.H, Seo, M.H, Park, K.W, Lee, W.K. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Domain-wise dissection of thermal stability enhancement in multidomain proteins.
Int.J.Biol.Macromol., 237, 2023
|
|
6L7Z
 
 | | Solution NMR structure of the N-terminal immunoglobulin variable domain of BTNL2 | | Descriptor: | Butyrophilin-like protein 2 | | Authors: | Basak, A.J, Lee, W, Samanta, D, De, S. | | Deposit date: | 2019-11-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into N-terminal IgV Domain of BTNL2, a T Cell Inhibitory Molecule, Suggests a Non-canonical Binding Interface for Its Putative Receptors.
J.Mol.Biol., 432, 2020
|
|
5VSO
 
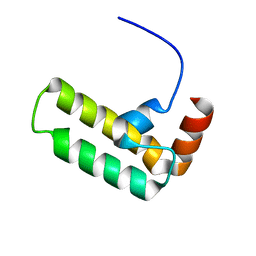 | | NMR structure of Ydj1 J-domain, a cytosolic Hsp40 from Saccharomyces cerevisiae | | Descriptor: | Yeast dnaJ protein 1 | | Authors: | Ciesielski, S.J, Tonelli, M, Lee, W, Cornilescu, G, Markley, J.L, Schilke, B.A, Ziegelhoffer, T, Craig, E.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Broadening the functionality of a J-protein/Hsp70 molecular chaperone system.
PLoS Genet., 13, 2017
|
|
2HQR
 
 | |
2HQO
 
 | |
2HQN
 
 | |
7WCG
 
 | |
