4N7V
 
 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep152 N-terminal fragment | | Descriptor: | Centrosomal protein of 152 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4N9J
 
 | | Crystal structure of the cryptic polo box domain of human Plk4 | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Ku, B, Kim, J.H, Lee, K.S, Kim, S.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4NQY
 
 | | The reduced form of MJ0499 | | Descriptor: | Isopropylmalate/citramalate isomerase large subunit, ZINC ION | | Authors: | Hwang, K.Y, Lee, E.H, Lee, K. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization and comparison of the large subunits of IPM isomerase and homoaconitase from Methanococcus jannaschii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2N8N
 
 | | Solution structure of translation initiation factor | | Descriptor: | Translation initiation factor IF-1 | | Authors: | Kim, D, Lee, K, Lee, B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics study of translation initiation factor 1 from Staphylococcus aureus suggests its RNA binding mode
Biochim.Biophys.Acta, 1865, 2017
|
|
4N7Z
 
 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep192 N-terminal fragment | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1DFW
 
 | | CONFORMATIONAL MAPPING OF THE N-TERMINAL SEGMENT OF SURFACTANT PROTEIN B IN LIPID USING 13C-ENHANCED FOURIER TRANSFORM INFRARED SPECTROSCOPY (FTIR) | | Descriptor: | LUNG SURFACTANT PROTEIN B | | Authors: | Gordon, L.M, Lee, K.Y.C, Lipp, M.M, Zasadzinski, J.A, Walther, F.J, Sherman, M.A, Waring, A.J. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | INFRARED SPECTROSCOPY | | Cite: | Conformational mapping of the N-terminal segment of surfactant protein B in lipid using 13C-enhanced Fourier transform infrared spectroscopy.
J.Pept.Res., 55, 2000
|
|
1P1A
 
 | | NMR structure of ubiquitin-like domain of hHR23B | | Descriptor: | UV excision repair protein RAD23 homolog B | | Authors: | Ryu, K.S, Lee, K.J, Bae, S.H, Kim, B.K, Kim, K.A, Choi, B.S. | | Deposit date: | 2003-04-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a
J.Biol.Chem., 278, 2003
|
|
5EXK
 
 | | Crystal structure of M. tuberculosis lipoyl synthase with 6-thiooctanoyl peptide intermediate | | Descriptor: | 5'-DEOXYADENOSINE, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1HA0
 
 | | HEMAGGLUTININ PRECURSOR HA0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HEMAGGLUTININ PRECURSOR), alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, J, Ho Lee, K, Steinhauer, D.A, Stevens, D.J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1998-10-08 | | Release date: | 1998-10-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation.
Cell(Cambridge,Mass.), 95, 1998
|
|
2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
8GRV
 
 | |
8GQQ
 
 | |
6O5H
 
 | | The effect of modifier structure on the activation of leukotriene A4 hydrolase aminopeptidase activity. | | Descriptor: | 4-{4-[(4-methoxyphenyl)methyl]phenyl}-1,3-thiazol-2-amine, Leukotriene A-4 hydrolase, ZINC ION | | Authors: | Noble, S.M, Lee, K.H, Paige, M. | | Deposit date: | 2019-03-03 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Modifier Structure on the Activation of Leukotriene A4Hydrolase Aminopeptidase Activity.
J.Med.Chem., 62, 2019
|
|
8IGI
 
 | | Crystal structure of HP1526 (XthA)- a base excision DNA repair protein in Helicobacter pylori | | Descriptor: | 1,3-BUTANEDIOL, Exodeoxyribonuclease (LexA), MANGANESE (II) ION | | Authors: | Dinh, T.T, Dao, O, Lee, K.H. | | Deposit date: | 2023-02-20 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease XthA (HP1526 protein) from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 663, 2023
|
|
3OBR
 
 | |
6J3L
 
 | | Solution structure of the N-terminal extended protuberant domain of eukaryotic ribosomal stalk protein P0 | | Descriptor: | 60S acidic ribosomal protein P0 | | Authors: | Choi, K.H.A, Lee, K.M, Yang, L, Wing-Heng Yu, C, Banfield, D.K, Ito, K, Uchiumi, T, Wong, K.B. | | Deposit date: | 2019-01-04 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mutagenesis Studies Evince the Role of the Extended Protuberant Domain of Ribosomal Protein uL10 in Protein Translation.
Biochemistry, 58, 2019
|
|
1KMR
 
 | |
3V64
 
 | | Crystal Structure of agrin and LRP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor-related protein 4, ... | | Authors: | Zong, Y, Zhang, B, Gu, S, Lee, K, Zhou, J, Yao, G, Figueiedo, D, Perry, K, Mei, L, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
3EF6
 
 | | Crystal structure of Toluene 2,3-Dioxygenase Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Friemann, R, Lee, K, Brown, E.N, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5J8H
 
 | | Structure of calmodulin in a complex with a peptide derived from a calmodulin-dependent kinase | | Descriptor: | CALCIUM ION, Calmodulin, Eukaryotic elongation factor 2 kinase | | Authors: | Alphonse, S, Lee, K, Piserchio, A, Tavares, C.D.J, Giles, D.H, Wellmann, R.M, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Eukaryotic Elongation Factor 2 Kinase by Calmodulin.
Structure, 24, 2016
|
|
2BD0
 
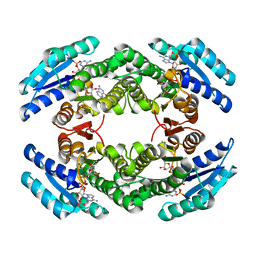 | | Chlorobium tepidum Sepiapterin Reductase complexed with NADP and Sepiapterin | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sepiapterin reductase | | Authors: | Supangat, S, Seo, K.H, Choi, Y.K, Park, Y.S, Lee, K.H. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of L-threo-tetrahydrobiopterin
J.Biol.Chem., 281, 2006
|
|
4MMN
 
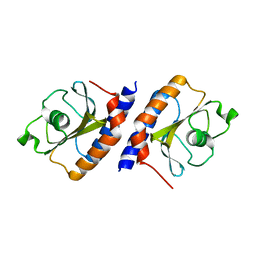 | | Structural and biochemical analysis of type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum | | Descriptor: | Putative uncharacterized protein Ta0848 | | Authors: | Kim, H.S, Kwak, G.H, Lee, K.T, Jo, C.H, Hwang, K.Y, Kim, H.Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum
Arch.Biochem.Biophys., 560, 2014
|
|
1Z9D
 
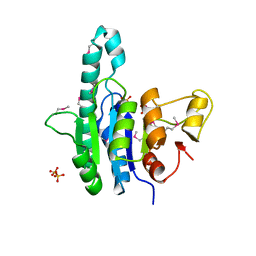 | | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes | | Descriptor: | SULFATE ION, uridylate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes
To be Published
|
|
4E7P
 
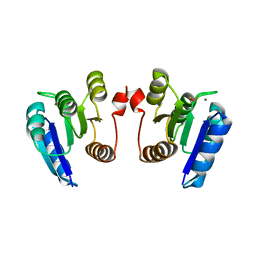 | | Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the presence of the phosphoryl analog beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator | | Authors: | Park, A.K, Moon, J.H, Lee, K.S, Chi, Y.M. | | Deposit date: | 2012-03-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the absence and presence of the phosphoryl analog beryllofluoride.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
6NDL
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
