5YOA
 
 | |
5YZU
 
 | |
2KI2
 
 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | Descriptor: | Ss-DNA binding protein 12RNP2 | | Authors: | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
1LX8
 
 | | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein | | Descriptor: | Excisionase | | Authors: | Sam, M.D, Papagiannis, C, Connolly, K.M, Corselli, L, Iwahara, J, Lee, J, Phillips, M, Wojciak, J.M, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2002-06-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein
J.Mol.Biol., 324, 2002
|
|
8SXR
 
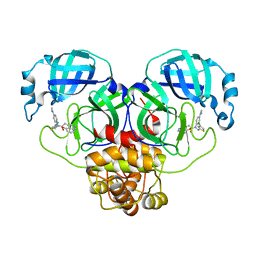 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | Descriptor: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2023-05-23 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
2KCC
 
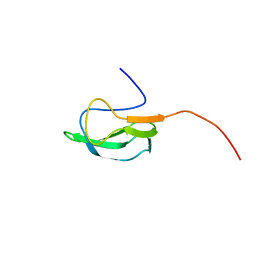 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
8CYU
 
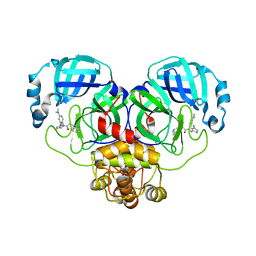 | | Crystal structure of SARS-CoV-2 Mpro with compound C5 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ7
 
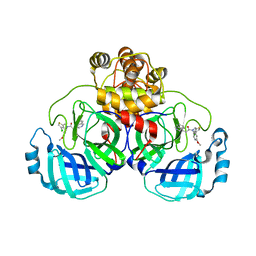 | | Crystal structure of SARS-CoV-2 Mpro with compound C2 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CYZ
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C4 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ4
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C3 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
2CW0
 
 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
2F0Y
 
 | |
4PKX
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
2PNL
 
 | | Crystal structure of VP4 protease from infectious pancreatic necrosis virus (IPNV) in space group P1 | | Descriptor: | GUANIDINE, Protease VP4 | | Authors: | Paetzel, M, Lee, J, Feldman, A.R, Delmas, B. | | Deposit date: | 2007-04-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the VP4 protease from infectious pancreatic necrosis virus reveals the acyl-enzyme complex for an intermolecular self-cleavage reaction.
J.Biol.Chem., 282, 2007
|
|
4PKE
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | PLATINUM (II) ION, Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-14 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
2PNM
 
 | |
1ZYR
 
 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
4FJS
 
 | | Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form | | Descriptor: | Ureidoglycolate dehydrogenase | | Authors: | Kim, M.I, Shin, I, Lee, J, Rhee, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
4MN0
 
 | | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state | | Descriptor: | Berovin, CALCIUM ION, MAGNESIUM ION | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Biochim.Biophys.Acta, 1834, 2013
|
|
7LKP
 
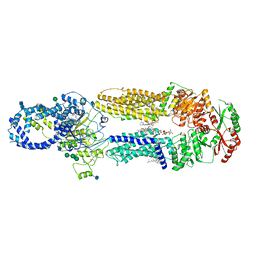 | | Structure of ATP-free human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
7LKZ
 
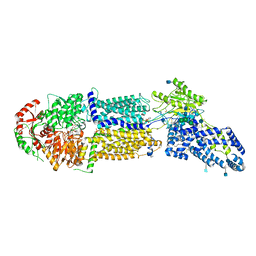 | | Structure of ATP-bound human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
4N1G
 
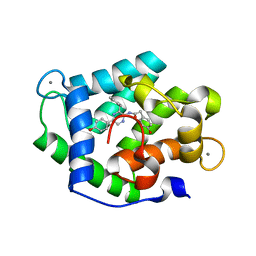 | | Crystal Structure of Ca(2+)- discharged F88Y obelin mutant from Obelia longissima at 1.50 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-10-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the F88Y obelin mutant before and after bioluminescence provide molecular insight into spectral tuning among hydromedusan photoproteins
Febs J., 281, 2014
|
|
3BA5
 
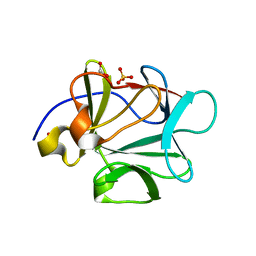 | |
3BAD
 
 | |
