3RAJ
 
 | |
5XG3
 
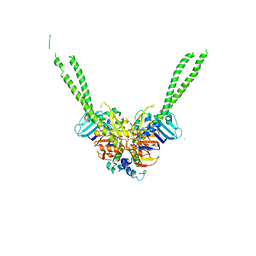 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
5XG2
 
 | |
6KHU
 
 | |
5YU3
 
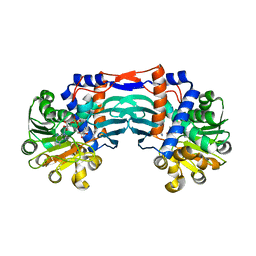 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROLINE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
6K3F
 
 | |
6KWA
 
 | |
2GMG
 
 | | Solution NMR Structure of protein PF0610 from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfG3 | | Descriptor: | hypothetical protein Pf0610 | | Authors: | Wang, X, Lee, H.S, Adams, M.W, Northeast Structural Genomics Consortium (NESG), Montelione, G.T, Prestegard, J.H. | | Deposit date: | 2006-04-06 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PF0610, a novel winged helix-turn-helix variant possessing a rubredoxin-like Zn ribbon motif from the hyperthermophilic archaeon, Pyrococcus furiosus.
Biochemistry, 46, 2007
|
|
3W9B
 
 | | Crystal structure of Candida antarctica lipase B with anion-tag | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Lipase B | | Authors: | Kim, S.K, Lee, H.H, Park, Y.C, Jeon, S.T, Son, S.H, Min, W.K, Seo, J.H. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anion tags improve extracellular production of Candida antarctica lipase B in Escherichia coli
To be Published
|
|
6KWB
 
 | |
6JRP
 
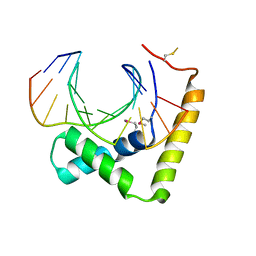 | | Crystal structure of CIC-HMG-ETV5-DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*AP*AP*TP*GP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Protein capicua homolog | | Authors: | Song, J.J, Lee, H. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of Capicua HMG-box domain complexed with the ETV5-DNA and its implications for Capicua-mediated cancers.
Febs J., 286, 2019
|
|
6JJK
 
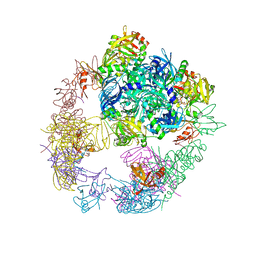 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
3WR5
 
 | |
2HEM
 
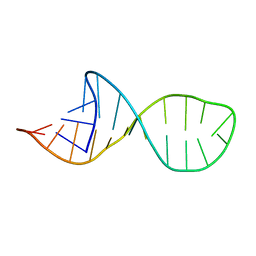 | | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit. | | Descriptor: | 5'-R(P*GP*GP*GP*AP*AP*GP*GP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*CP*GP*GP*CP*CP*C)-3' | | Authors: | Zhao, Q, Nagaswamy, U, Lee, H, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2006-06-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit
Nucleic Acids Res., 33, 2005
|
|
2KW5
 
 | | Solution NMR Structure of the Slr1183 protein from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Rossi, P, Forouhar, F, Lee, H, Lange, O, Mao, B, Lemak, A, Maglaqui, M, Belote, R, Ciccosanti, C, Foote, E, Sahdev, S, Acton, T, Xiao, R, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KJ5
 
 | | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C | | Descriptor: | Phage integrase | | Authors: | Mills, J.L, Eletsky, A, Ghosh, A, Wang, D, Lee, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C.
To be Published
|
|
2KD1
 
 | | Solution NMR structure of the integrase-like domain from Bacillus cereus ordered locus BC_1272. Northeast Structural Genomics Consortium Target BcR268F | | Descriptor: | DNA integration/recombination/invertion protein | | Authors: | Rossi, P, Lee, H, Maglaqui, M, Foote, E.L, Buchwald, W.A, Jiang, M, Swapna, G.V.T, Nair, R, Xiao, R, Acton, T.B, Rost, B, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Integrase-Like Domain from Bacillus cereus Ordered Locus BC_1272. Northeast Structural Genomics Consortium Target BcR268F.
To be Published
|
|
3V5K
 
 | | HLA2.1 KVAELVWFL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HIV-based altered-peptide ligand KVAELVWFL, ... | | Authors: | Collins, E.J, Lee, H.Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Prediction of Immunogenicity of altered-peptide ligands to HIV bound to MHC
To be Published
|
|
2L7K
 
 | | Solution NMR Structure of protein CD1104.2 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130 | | Descriptor: | Uncharacterized protein | | Authors: | Pulavarti, S.V.S.R.K, Eletsky, A, Mills, J.L, Sukumaran, D.K, Wang, H, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-13 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CfR130 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130
To be Published
|
|
3V5H
 
 | | HLA-A2.1 KVAEIVHFL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HIV-based altered-peptide ligand KVAEIVHFL, ... | | Authors: | Collins, E.J, Lee, H.Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Prediction of immunogenicity of altered-peptide ligands for HIV therapy
To be Published
|
|
2HNV
 
 | | Crystal Structure of a Dipeptide Complex of the Q58V Mutant of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2I66
 
 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5S)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
5Z2W
 
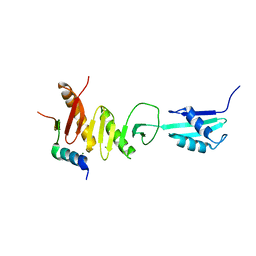 | | Crystal structure of the bacterial cell division protein FtsQ and FtsB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsQ, MAGNESIUM ION | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the FtsQ/FtsB/FtsL Complex, a Key Component of the Divisome.
Sci Rep, 8, 2018
|
|
3V5D
 
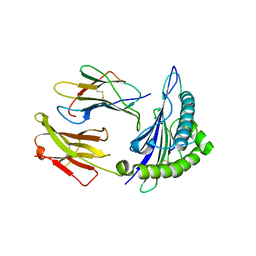 | | HLA-A2.1 KVAELVHFL | | Descriptor: | Beta-2-microglobulin, HIV peptide KVAELVHFL, HLA class I histocompatibility antigen, ... | | Authors: | Collins, E.J, Lee, H.Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of conformation and immunogenicity of peptides bound to MHC molecules
To be Published
|
|
3UVN
 
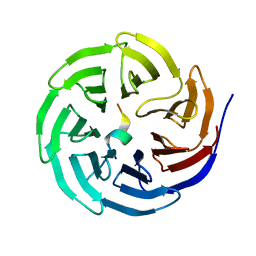 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
