7CH3
 
 | | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) triple mutant (K264A, E265A, K266A) | | Descriptor: | L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Cao, T.P, Dhanasingh, I, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-07-04 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) triple mutant (K264A, E265A, K266A)
To Be Published
|
|
2WXP
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with GDC-0941. | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXG
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with SW13. | | Descriptor: | 2-{[4-amino-3-(3-fluoro-5-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
1YZS
 
 | |
1BXB
 
 | | XYLOSE ISOMERASE FROM THERMUS THERMOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
6PC5
 
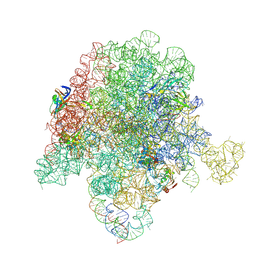 | | E. coli 50S ribosome bound to compounds 46 and VS1 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
1NME
 
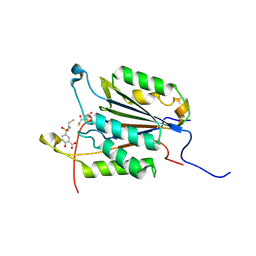 | | Structure of Casp-3 with tethered salicylate | | Descriptor: | 2-HYDROXY-5-(2-MERCAPTO-ETHYLSULFAMOYL)-BENZOIC ACID, 3-(2-MERCAPTO-ACETYLAMINO)-4-OXO-PENTANOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, B, DeLano, W, Choong, I.C, Flanagan, M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
3LMC
 
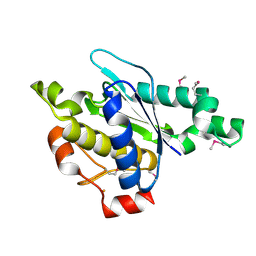 | | Crystal Structure of zinc-dependent peptidase from Methanocorpusculum labreanum (strain Z), Northeast Structural Genomics Consortium Target MuR16 | | Descriptor: | FE (III) ION, Peptidase, zinc-dependent, ... | | Authors: | Kuzin, A, Ashok, S, Vorobiev, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Northeast Structural Genomics Consortium Target MuR16
To be published
|
|
5YSZ
 
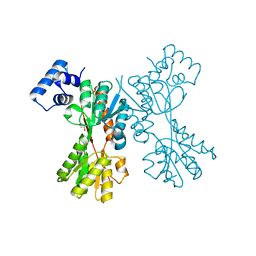 | | transcriptional regulator CelR-cellobiose complex | | Descriptor: | GLYCEROL, Transcriptional regulator, LacI family, ... | | Authors: | Fu, Y, Yeom, S.Y, Lee, D.H, Lee, S.G. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structural and functional analyses of the cellulase transcription regulator CelR
FEBS Lett., 592, 2018
|
|
3LMF
 
 | | Crystal Structure of Nmul_A1745 protein from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR72 | | Descriptor: | Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target NmR72
To be Published
|
|
3MEJ
 
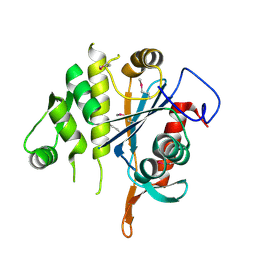 | | Crystal structure of putative transcriptional regulator ywtF from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR736 | | Descriptor: | CHLORIDE ION, transcriptional regulator ywtF | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR736
To be Published
|
|
3F2Z
 
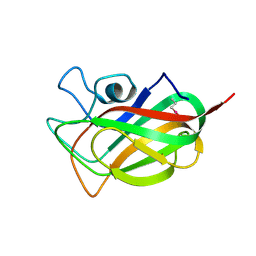 | | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B | | Descriptor: | uncharacterized protein BF3579 | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, Foote, E.L, Ciccosanti, C, Lee, D, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B
To be Published
|
|
1TAQ
 
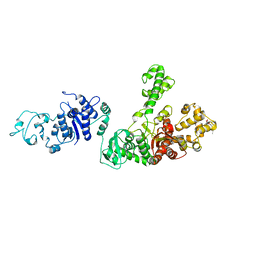 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
3JSR
 
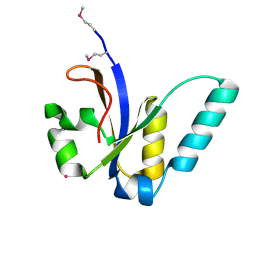 | | X-Ray structure of All0216 protein from Nostoc sp. PCC 7120 at the resolution 1.8A. Northeast Structural Genomics Consortium target NsR236 | | Descriptor: | All0216 protein, POTASSIUM ION | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray structure of All0216 protein from Nostoc sp. PCC 7120 at the resolution 1.8A
To be Published
|
|
7CYY
 
 | | Crystal structure of Arabinose isomerase from hybrid AI8 with Adonitol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-ribitol, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic hybrid AI8 with Adonitol
To Be Published
|
|
1V9Y
 
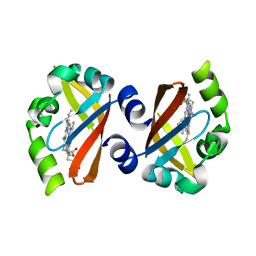 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
7CHL
 
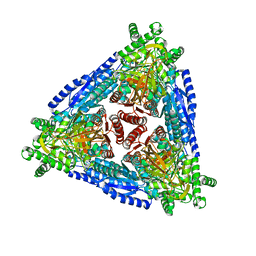 | | Crystal structure of hybrid Arabinose isomerase AI-10 | | Descriptor: | Hybrid Arabinose isomerase, MANGANESE (II) ION, SODIUM ION | | Authors: | Cao, T.P, Dhanasingh, I, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of hybrid Arabinose isomerase AI-10
To Be Published
|
|
1UPS
 
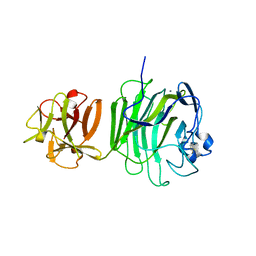 | | GlcNAc[alpha]1-4Gal releasing endo-[beta]-galactosidase from Clostridium perfringens | | Descriptor: | CALCIUM ION, GLCNAC-ALPHA-1,4-GAL-RELEASING ENDO-BETA-GALACTOSIDASE | | Authors: | Tempel, W, Liu, Z.-J, Horanyi, P.S, Deng, L, Lee, D, Newton, M.G, Rose, J.P, Ashida, H, Li, S.-C, Li, Y.-T, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-10-10 | | Release date: | 2004-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Three-dimensional structure of GlcNAcalpha1-4Gal releasing endo-beta-galactosidase from Clostridium perfringens.
Proteins, 59, 2005
|
|
2PK8
 
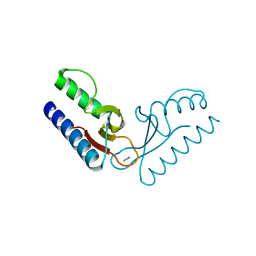 | | Crystal structure of an uncharacterized protein PF0899 from Pyrococcus furiosus | | Descriptor: | GOLD (I) CYANIDE ION, Uncharacterized protein PF0899 | | Authors: | Liu, Z.J, Tempel, W, Chen, L, Shah, A, Lee, D, Clancy-Kelley, L.L, Dillard, B.D, Rose, J.P, Sugar, F.J, Jenny Jr, F.E, Lee, H.S, Izumi, M, Shah, C, Poole III, F.L, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the hypothetical protein PF0899 from Pyrococcus furiosus at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2KC7
 
 | | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218 | | Descriptor: | bfr218_protein | | Authors: | Eletsky, A, Wu, Y, Sukumaran, D, Lee, H, Lee, D.Y, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-17 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218
To be Published
|
|
3HZE
 
 | | Crystal Structure of Ycf54 protein from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR59 | | Descriptor: | PHOSPHATE ION, Ycf54 protein | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target TeR59
To be Published
|
|
3IDU
 
 | | Crystal Structure of the CARDB domain of the PF1109 protein in complex with di-metal ions from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR193A | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target PfR193A
To be Published
|
|
7ER9
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Febuxostat (TEI) | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ER7
 
 | | Crystal structure of hyman Biliverdin IX-beta reductase B with Tamibarotene (A80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ER6
 
 | | Crystal structure of human Biliverdin IX-beta reductase B | | Descriptor: | Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
