7U17
 
 | | TMEM106B(120-254) T185S singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type B case 2 (case 7). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U0Z
 
 | | High-resolution map of tau filament from progressive supranuclear palsy (PSP) case 1 | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau | | Authors: | FItzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U10
 
 | | TMEM106B(120-254) protofilament from progressive supranuclear palsy (PSP) case 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U14
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type C (case 8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U11
 
 | | TMEM106B(120-254) protofilament from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U12
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U13
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
4KSD
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A, R2 protein | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (4.1001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OQ3
 
 | |
2ZY5
 
 | | R487A mutant of L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2ZY2
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate 4-carboxylyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2ZY3
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
5Z26
 
 | |
1LM4
 
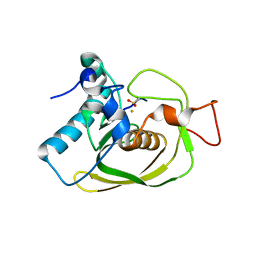 | | Structure of Peptide Deformylase from Staphylococcus aureus at 1.45 A | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase PDF1 | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LME
 
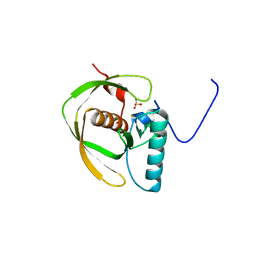 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
5E1J
 
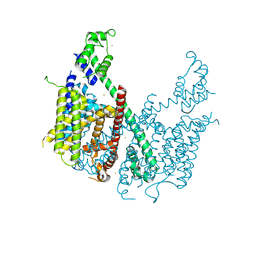 | | Structure of voltage-gated two-pore channel TPC1 from Arabidopsis thaliana | | Descriptor: | BARIUM ION, CALCIUM ION, Two pore calcium channel protein 1 | | Authors: | Guo, J, Zeng, W, Chen, Q, Lee, C, Chen, L, Yang, Y, Jiang, Y. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.308 Å) | | Cite: | Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana.
Nature, 531, 2016
|
|
2M30
 
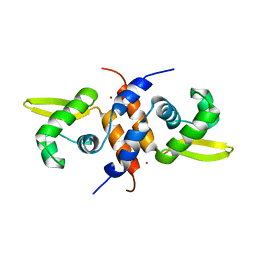 | | Solution NMR refinement of a metal ion bound protein using quantum mechanical/molecular mechanical and molecular dynamics methods | | Descriptor: | Repressor protein, ZINC ION | | Authors: | Chakravorty, D.K, Wang, B.I, Lee, C.I, Guerra, A.J, Giedroc, D.P, Merz Jr, K.M, Arunkumar, A.I, Pennella, M, Kong, X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR refinement of a metal ion bound protein using metal ion inclusive restrained molecular dynamics methods.
J.Biomol.Nmr, 56, 2013
|
|
1LM6
 
 | | Crystal Structure of Peptide Deformylase from Streptococcus pneumoniae | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase DEFB | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
2ZY4
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | CHLORIDE ION, L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
1N5N
 
 | | Crystal Structure of Peptide Deformylase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Peptide deformylase, ZINC ION | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
5GYD
 
 | | Crystal Structure of Mdm12 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Mitochondrial distribution and morphology protein 12 | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2016-09-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex
EMBO Rep., 17, 2016
|
|
1XAK
 
 | |
7VIZ
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (250 ns time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
