7B7D
 
 | | Yeast 80S ribosome bound to eEF3 and A/A- and P/P-tRNAs | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Ranjan, N, Pochopien, A.A, Wu, C.C, Beckert, B, Blanchet, S, Green, R, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | eEF3 promotes late stages of tRNA translocation including E-tRNA release from the ribosome
To Be Published
|
|
1GWX
 
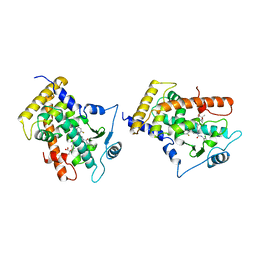 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 2-(4-{3-[1-[2-(2-CHLORO-6-FLUORO-PHENYL)-ETHYL]-3-(2,3-DICHLORO-PHENYL)-UREIDO]-PROPYL}-PHENOXY)-2-METHYL-PROPIONIC ACID, PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-17 | | Release date: | 2000-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
1YZ6
 
 | |
3OOV
 
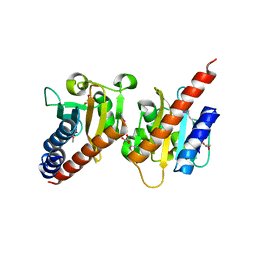 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
4GVV
 
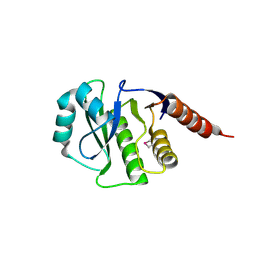 | | Crystal Structure of de novo design serine hydrolase OSH55.27, Northeast Structural Genomics Consortium (NESG) Target OR246 | | Descriptor: | De novo design serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR246
To be Published
|
|
1DQU
 
 | | CRYSTAL STRUCTURE OF THE ISOCITRATE LYASE FROM ASPERGILLUS NIDULANS | | Descriptor: | ISOCITRATE LYASE | | Authors: | Britton, K.L, Langridge, S.J, Baker, P.J, Weeradechapon, K, Sedelnikova, S.E, De Lucas, J.R, Rice, D.W, Turner, G. | | Deposit date: | 2000-01-05 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure and active site location of isocitrate lyase from the fungus Aspergillus nidulans.
Structure Fold.Des., 8, 2000
|
|
8FZQ
 
 | | Dehosphorylated, ATP-bound human cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Levring, J, Terry, D.S, Kilic, Z, Fitzgerald, G.A, Blanchard, S.C, Chen, J. | | Deposit date: | 2023-01-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CFTR function, pathology and pharmacology at single-molecule resolution.
Nature, 616, 2023
|
|
3RXY
 
 | | Crystal structure of NIF3 superfamily protein from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Michalska, K, Tesar, C, Clancy, S, Otwinowski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-10 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NIF3 superfamily protein from Sphaerobacter thermophilus
To be Published
|
|
3JR7
 
 | | The crystal structure of the protein of DegV family COG1307 with unknown function from Ruminococcus gnavus ATCC 29149 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the protein of DegV family COG1307 with unknown function from Ruminococcus gnavus ATCC 29149
To be Published
|
|
3OOO
 
 | | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V | | Descriptor: | Proline dipeptidase | | Authors: | Fan, Y, Wu, R, Morales, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V
To be Published
|
|
6GZ3
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-1 (TI-POST-1) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
3M4R
 
 | | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-11 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum
TO BE PUBLISHED
|
|
5WL2
 
 | | VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Descriptor: | Germline-reverted light chain of CR9114, Heavy chain of VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem.
To Be Published
|
|
4MTN
 
 | |
4NAS
 
 | | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446.
To be Published
|
|
8FAK
 
 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | Descriptor: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
5WKO
 
 | |
6V5M
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
4JGK
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275 | | Descriptor: | evolved variant of a designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275
To be Published
|
|
3HYY
 
 | | Crystal structure of Hsp90 with fragment 37-D04 | | Descriptor: | Heat shock protein HSP 90-alpha, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment 37-D04
TO BE PUBLISHED
|
|
6N04
 
 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
3TT2
 
 | | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GCN5-related N-acetyltransferase, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus
To be Published
|
|
1G24
 
 | |
4KTB
 
 | | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Tan, K, Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603
To be Published
|
|
3HZ1
 
 | | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03
TO BE PUBLISHED
|
|
