2R78
 
 | |
2GNP
 
 | | Structural Genomics, the crystal structure of a transcriptional regulator from Streptococcus pneumoniae TIGR4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, transcriptional regulator | | Authors: | Tan, K, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a transcriptional regulator from Streptococcus pneumoniae
TIGR4
To be Published
|
|
3TP4
 
 | | Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
2GS5
 
 | |
2OCZ
 
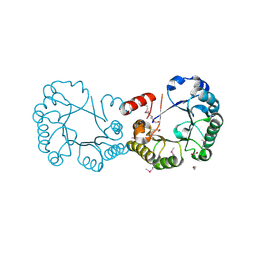 | | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes. | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase, MAGNESIUM ION | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes.
TO BE PUBLISHED
|
|
1IN6
 
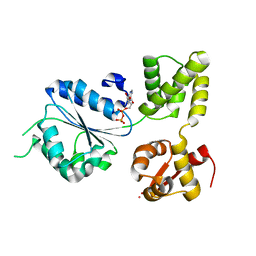 | | THERMOTOGA MARITIMA RUVB K64R MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN7
 
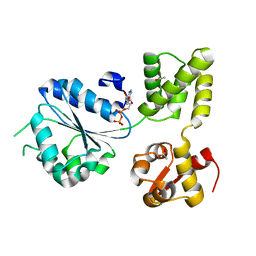 | | THERMOTOGA MARITIMA RUVB R170A | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
3QOO
 
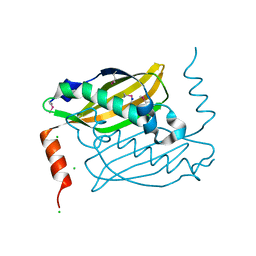 | | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans
TO BE PUBLISHED
|
|
1JKE
 
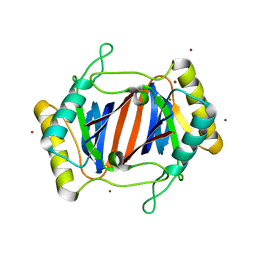 | | D-Tyr tRNATyr deacylase from Escherichia coli | | Descriptor: | D-Tyr-tRNATyr deacylase, ZINC ION | | Authors: | Ferri-Fioni, M.L, Schmitt, E, Soutourina, J, Plateau, P, Mechulam, Y, Blanquet, S. | | Deposit date: | 2001-07-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of crystalline D-Tyr-tRNA(Tyr) deacylase. A representative of a new class of tRNA-dependent hydrolases.
J.Biol.Chem., 276, 2001
|
|
3RPC
 
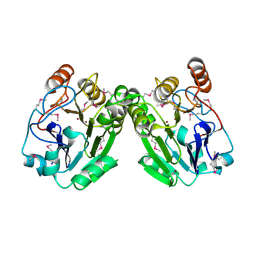 | |
3R6D
 
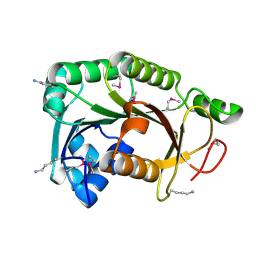 | |
2RK5
 
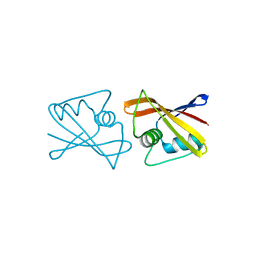 | |
1XTY
 
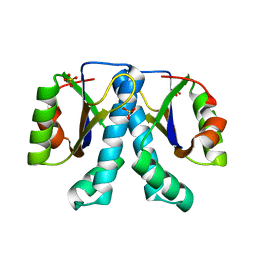 | | Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Fromant, M, Schmitt, E, Mechulam, Y, Lazennec, C, Plateau, P, Blanquet, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and identification of active site residues of Sulfolobus solfataricus peptidyl-tRNA hydrolase.
Biochemistry, 44, 2005
|
|
2HNG
 
 | | The Crystal Structure of Protein of Unknown Function SP1558 from Streptococcus pneumoniae | | Descriptor: | Hypothetical protein | | Authors: | Kim, Y, Zhang, D, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-12 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Crystal Structure of Hypothetical Protein SP_1558 from Streptococcus pneumoniae
To be Published, 2006
|
|
2I5E
 
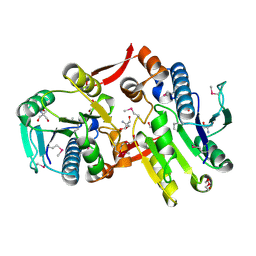 | | Crystal Structure of a Protein of Unknown Function MM2497 from Methanosarcina mazei Go1, Probable Nucleotidyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Hypothetical protein MM_2497 | | Authors: | Tan, K, Du, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a hypothetical protein MM_2497 from Methanosarcina mazei Go1
To be Published
|
|
3TOV
 
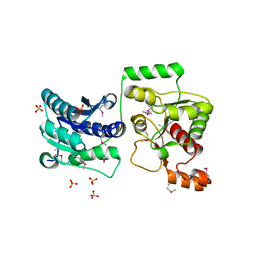 | |
4Y0D
 
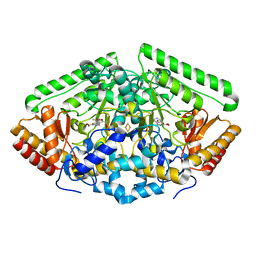 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
3L0Z
 
 | |
3L5Z
 
 | | Crystal structure of transcriptional regulator, GntR family from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Transcriptional regulator, ... | | Authors: | Chang, C, Hatzos, C, Feldmann, B, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of transcriptional regulator, GntR family from Bacillus cereus
To be Published
|
|
3LHE
 
 | | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne | | Descriptor: | CHLORIDE ION, GLYCEROL, GntR family Transcriptional regulator | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne
To be Published
|
|
3LLV
 
 | | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A | | Descriptor: | Exopolyphosphatase-related protein, PHOSPHATE ION | | Authors: | Stein, A.J, Chang, C, Weger, A, Hendricks, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A
To be Published
|
|
4Y0H
 
 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | 4-aminobutyrate aminotransferase, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-06 | | Release date: | 2015-03-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
3LAE
 
 | | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, UPF0053 protein HI0107, ... | | Authors: | Tan, K, Li, H, Bargassa, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20
To be Published
|
|
3LF0
 
 | | Crystal structure of the ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory protein P-II | | Authors: | Shetty, N.D, Palaninathan, S.K, Reddy, M.C.M, Owen, J.L, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-15 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein.
Protein Sci., 19, 2010
|
|
3LSG
 
 | |
