1KTR
 
 | | Crystal Structure of the Anti-His Tag Antibody 3D5 Single-Chain Fragment (scFv) in Complex with a Oligohistidine peptide | | Descriptor: | Anti-his tag antibody 3d5 variable light chain, Peptide linker, Anti-his tag antibody 3d5 variable heavy chain, ... | | Authors: | Kaufmann, M, Lindner, P, Honegger, A, Blank, K, Tschopp, M, Capitani, G, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-01-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the anti-His tag antibody 3D5 single-chain fragment complexed to its antigen.
J.Mol.Biol., 318, 2002
|
|
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6NFK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
6YX5
 
 | | Structure of DrrA from Legionella pneumophilia in complex with human Rab8a | | Descriptor: | MAGNESIUM ION, Multifunctional virulence effector protein DrrA, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Schneider, S, Du, J, von Wrisberg, M.K, Lang, K, Itzen, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1.
Nat Commun, 12, 2021
|
|
6BQH
 
 | | Crystal structure of 5-HT2C in complex with ritanserin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, 6-(2-{4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl}ethyl)-7-methyl-5H-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ... | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
1Z7D
 
 | | Ornithine aminotransferase PY00104 from Plasmodium Yoelii | | Descriptor: | ornithine aminotransferase | | Authors: | Walker, J.R, Alam, Z, Amani, M, Lew, J, Wasney, G, Boulanger, K, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-24 | | Release date: | 2005-07-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2LHU
 
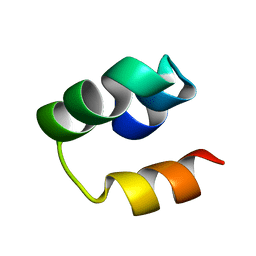 | | Structural Insight into the Unique Cardiac Myosin Binding Protein-C Motif: A Partially Folded Domain | | Descriptor: | Mybpc3 protein | | Authors: | Howarth, J.W, Rosevear, P.R, Ramisetti, S, Nolan, K, Sadayappan, S. | | Deposit date: | 2011-08-18 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Unique Cardiac Myosin-binding Protein-C Motif: A PARTIALLY FOLDED DOMAIN.
J.Biol.Chem., 287, 2012
|
|
5WOF
 
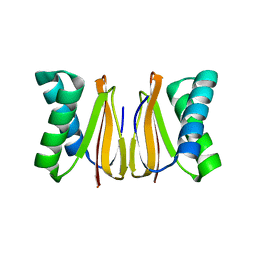 | | 1.65 ANGSTROM STRUCTURE OF THE DYNEIN LIGHT CHAIN 1 FROM PLASMODIUM FALCIPARUM | | Descriptor: | Dynein light chain 1, putative | | Authors: | Walker, J.R, Lew, J, Amani, M, Alam, Z, Wasney, G, Boulanger, K, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Botchkarev, A, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genome-scale Protein Expression and Structural Biology of
Plasmodium Falciparum and Related Apicomplexan Organisms.
MOL.BIOCHEM.PARASITOL., 151, 2007
|
|
2OZ0
 
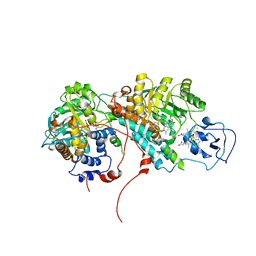 | | Mechanistic and Structural Studies of H373Q Flavocytochrome b2: Effects of Mutating the Active Site Base | | Descriptor: | Cytochrome b2, FLAVIN MONONUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tsai, C.-L, Gokulan, K, Sobrado, P, Sacchettini, J.C, Fitzpatrick, P.F. | | Deposit date: | 2007-02-23 | | Release date: | 2007-09-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of H373Q flavocytochrome b2: effects of mutating the active site base.
Biochemistry, 46, 2007
|
|
1WQJ
 
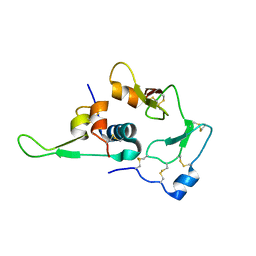 | | Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor binding protein 4 | | Authors: | Siwanowicz, I, Popowicz, G.M, Wisniewska, M, Huber, R, Kuenkele, K.P, Lang, K, Engh, R.A, Holak, T.A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Structure, 13, 2005
|
|
7N44
 
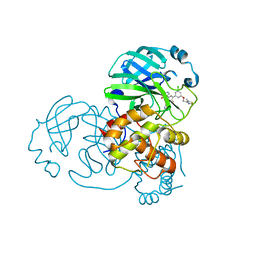 | | Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13) | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Reilly, R.A, Zhang, C.H, Deshmukh, M.G, Ippolito, J.A, Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency.
Acs Med.Chem.Lett., 12, 2021
|
|
1YNV
 
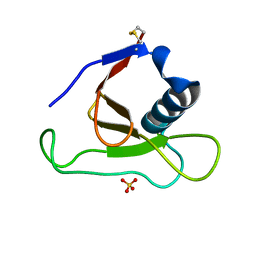 | | Asp79 makes a large, unfavorable contribution to the stability of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Trevino, S.R, Gokulan, K, Newsom, S, Thurlkill, R.L, Shaw, K.L, Mitkevich, V.A, Makarov, A.A, Sacchettini, J.C, Scholtz, J.M, Pace, C.N. | | Deposit date: | 2005-01-25 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Asp79 Makes a Large, Unfavorable Contribution to the Stability of RNase Sa.
J.Mol.Biol., 354, 2005
|
|
5TVN
 
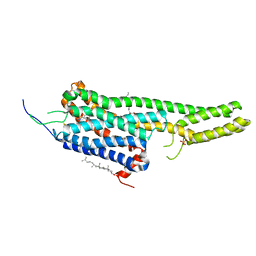 | | Crystal structure of the LSD-bound 5-HT2B receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, S, McCorvy, J.D, Betz, R.M, Venkatakrishnan, A.J, Levit, A, Lansu, K, Schools, Z.L, Che, T, Nichols, D.E, Shoichet, B.K, Dror, R.O, Roth, B.L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of an LSD-Bound Human Serotonin Receptor.
Cell, 168, 2017
|
|
6UVU
 
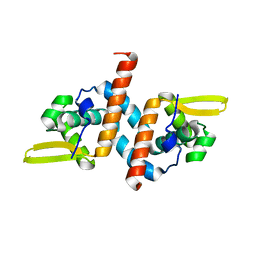 | | Crystal structure of the AntR antimony-specific transcriptional repressor | | Descriptor: | ArsR family transcriptional regulator | | Authors: | Thiruselvam, V, Banumathi, S, Palani, K, Manohar, R, Rosen, B.P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of AntR, an Sb(III) responsive transcriptional repressor.
Mol.Microbiol., 116, 2021
|
|
6STG
 
 | | Human Rab8a phosphorylated at Ser111 in complex with GPPNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vieweg, S, Mulholland, K, Braeuning, B, Kachariya, N, Lai, Y, Toth, R, Sattler, M, Groll, M, Itzen, A, Muqit, M.M.K. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PINK1-dependent phosphorylation of Serine111 within the SF3 motif of Rab GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72.
Biochem.J., 477, 2020
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRY
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6STF
 
 | | Human Rab8a phosphorylated at Ser111 in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-8A | | Authors: | Vieweg, S, Mulholland, K, Braeuning, B, Kachariya, N, Lai, Y, Toth, R, Sattler, M, Groll, M, Itzen, A, Muqit, M.M.K. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PINK1-dependent phosphorylation of Serine111 within the SF3 motif of Rab GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72.
Biochem.J., 477, 2020
|
|
3EII
 
 | | Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-16 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
3EJA
 
 | | Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
3C3Z
 
 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to ribostamycin | | Descriptor: | HIV-1 subtype F genomic RNA, RIBOSTAMYCIN | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
2R80
 
 | | Pigeon Hemoglobin (OXY form) | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Ponnuswamy, M.N, Packianathan, C, Sundaresan, S, Neelagandan, K, Palani, K, Muller, J.J, Heinemann, U. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-ray crystal structure analysis of Hemolgobin from Pigeon (Columba Livia) at 1.44 angstrom
To be Published
|
|
