1PDU
 
 | | Ligand-binding domain of Drosophila orphan nuclear receptor DHR38 | | Descriptor: | nuclear hormone receptor HR38 | | Authors: | Baker, K.D, Shewchuk, L.M, Korlova, T, Makishima, M, Hassell, A.M, Wisely, B, Caravella, J.A, Lambert, M.H, Wilson, T.M, Mangelsdorf, D.J. | | Deposit date: | 2003-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway.
Cell(Cambridge,Mass.), 113, 2003
|
|
2FG7
 
 | | N-succinyl-L-ornithine transcarbamylase from B. fragilis complexed with carbamoyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, SULFATE ION, ... | | Authors: | Shi, D, Yu, X, Malamy, M.H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
J.Biol.Chem., 281, 2006
|
|
1X78
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1, [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1X7B
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
2A1H
 
 | | X-ray crystal structure of human mitochondrial branched chain aminotransferase (BCATm) complexed with gabapentin | | Descriptor: | ACETIC ACID, PYRIDOXAL-5'-PHOSPHATE, [1-(AMINOMETHYL)CYCLOHEXYL]ACETIC ACID, ... | | Authors: | Goto, M, Miyahara, I, Hirotsu, K, Conway, M, Yennawar, N, Islam, M.M, Hutson, S.M. | | Deposit date: | 2005-06-20 | | Release date: | 2005-09-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin.
J.Biol.Chem., 280, 2005
|
|
1X76
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-697 | | Descriptor: | 5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-CARBONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1X7E
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR ALPHA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor 1 (alpha), [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE, steroid receptor coactivator-3 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
3DZD
 
 | | Crystal structure of sigma54 activator NTRC4 in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SODIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-07-29 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
2FG6
 
 | | N-succinyl-L-ornithine transcarbamylase from B. fragilis complexed with sulfate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, SULFATE ION, putative ornithine carbamoyltransferase | | Authors: | Shi, D, Yu, X, Malamy, M.H, Allewell, N.M, Mendel, T. | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
J.Biol.Chem., 281, 2006
|
|
3E7L
 
 | | Crystal structure of sigma54 activator NtrC4's DNA binding domain | | Descriptor: | Transcriptional regulator (NtrC family), ZINC ION | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
2GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
3VTT
 
 | |
1FM9
 
 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND GI262570 AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA, ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
1FM6
 
 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND ROSIGLITAZONE AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
4R0S
 
 | | Crystal structure of P. aeruginosa TpbA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
7AH1
 
 | | L19 diabody fragment from immunocytokine L19-IL2 | | Descriptor: | Anti-(ED-B) scFV | | Authors: | Ongaro, T, Guarino, S.R, Scietti, L, Palamini, M, Wulhfard, S, Villa, A, Neri, D, Forneris, F. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
J.Struct.Biol., 213, 2021
|
|
7AI7
 
 | | MutS in Intermediate state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*CP*TP*TP*AP*GP*CP*TP*TP*AP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*TP*AP*AP*CP*TP*AP*AP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AIB
 
 | | MutS-MutL in clamp state | | Descriptor: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI5
 
 | | MutS in Scanning state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*AP*AP*TP*TP*CP*GP*CP*CP*CP*TP*AP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*TP*AP*GP*GP*GP*CP*GP*AP*AP*TP*TP*GP*GP*GP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI6
 
 | | MutS in mismatch bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein MutS | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AIC
 
 | | MutS-MutL in clamp state (kinked clamp domain) | | Descriptor: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3SFI
 
 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 5,5,5-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}pentan-1-one, Transcriptional regulatory repressor protein (TETR-family) | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
4ZHV
 
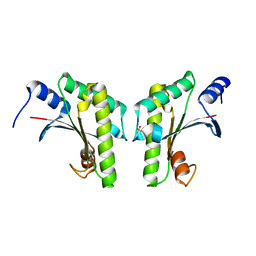 | | Crystal structure of a bacterial signalling protein | | Descriptor: | SULFATE ION, YfiB | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
5YKR
 
 | |
5YKT
 
 | |
